CellSpecks: A Software for Automated Detection and Analysis of Calcium Channels in Live Cells
- PMID: 30447989
- PMCID: PMC6289074
- DOI: 10.1016/j.bpj.2018.10.015
CellSpecks: A Software for Automated Detection and Analysis of Calcium Channels in Live Cells
Abstract
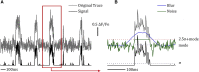
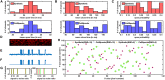
To couple the fidelity of patch-clamp recording with a more high-throughput screening capability, we pioneered a, to our knowledge, novel approach to single-channel recording that we named "optical patch clamp." By using highly sensitive fluorescent Ca2+ indicator dyes in conjunction with total internal fluorescence microscopy techniques, we monitor Ca2+ flux through individual Ca2+-permeable channels. This approach provides information about channel gating analogous to patch-clamp recording at a time resolution of ∼2 ms with the additional advantage of being massively parallel, providing simultaneous and independent recording from thousands of channels in the native environment. However, manual analysis of the data generated by this technique presents severe challenges because a video recording can include many thousands of frames. To overcome this bottleneck, we developed an image processing and analysis framework called CellSpecks capable of detecting and fully analyzing the kinetics of ion channels within a video sequence. By using randomly generated synthetic data, we tested the ability of CellSpecks to rapidly and efficiently detect and analyze the activity of thousands of ion channels, including openings for a few milliseconds. Here, we report the use of CellSpecks for the analysis of experimental data acquired by imaging muscle nicotinic acetylcholine receptors and the Alzheimer's disease-associated amyloid β pores with multiconductance levels in the plasma membrane of Xenopus laevis oocytes. We show that CellSpecks can accurately and efficiently generate location maps and create raw and processed fluorescence time traces; histograms of mean open times, mean close times, open probabilities, durations, and maximal amplitudes; and a "channel chip" showing the activity of all channels as a function of time. Although we specifically illustrate the application of CellSpecks for analyzing data from Ca2+ channels, it can be easily customized to analyze other spatially and temporally localized signals.
Copyright © 2018 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures




Similar articles
-
"Optical patch-clamping": single-channel recording by imaging Ca2+ flux through individual muscle acetylcholine receptor channels.J Gen Physiol. 2005 Sep;126(3):179-92. doi: 10.1085/jgp.200509331. Epub 2005 Aug 15. J Gen Physiol. 2005. PMID: 16103278 Free PMC article.
-
Imaging single-channel calcium microdomains.Cell Calcium. 2006 Nov-Dec;40(5-6):413-22. doi: 10.1016/j.ceca.2006.08.006. Epub 2006 Oct 25. Cell Calcium. 2006. PMID: 17067668 Free PMC article. Review.
-
Analyzing and Modeling the Kinetics of Amyloid Beta Pores Associated with Alzheimer's Disease Pathology.PLoS One. 2015 Sep 8;10(9):e0137357. doi: 10.1371/journal.pone.0137357. eCollection 2015. PLoS One. 2015. PMID: 26348728 Free PMC article.
-
Imaging the activity and localization of single voltage-gated Ca(2+) channels by total internal reflection fluorescence microscopy.Biophys J. 2004 May;86(5):3250-9. doi: 10.1016/S0006-3495(04)74373-8. Biophys J. 2004. PMID: 15111438 Free PMC article.
-
Optical single-channel recording: imaging Ca2+ flux through individual ion channels with high temporal and spatial resolution.J Biomed Opt. 2005 Jan-Feb;10(1):11002. doi: 10.1117/1.1846074. J Biomed Opt. 2005. PMID: 15847568 Review.
Cited by
-
Noise analysis of cytosolic calcium image data.Cell Calcium. 2020 Mar;86:102152. doi: 10.1016/j.ceca.2019.102152. Epub 2019 Dec 18. Cell Calcium. 2020. PMID: 31918030 Free PMC article.
-
PunctaSpecks: A tool for automated detection, tracking, and analysis of multiple types of fluorescently labeled biomolecules.Cell Calcium. 2020 Jul;89:102224. doi: 10.1016/j.ceca.2020.102224. Epub 2020 May 25. Cell Calcium. 2020. PMID: 32502904 Free PMC article.
-
From Seeing to Simulating: A Survey of Imaging Techniques and Spatially-Resolved Data for Developing Multiscale Computational Models of Liver Regeneration.Front Syst Biol. 2022;2:917191. doi: 10.3389/fsysb.2022.917191. Epub 2022 Jun 6. Front Syst Biol. 2022. PMID: 37575468 Free PMC article.
References
-
- Bootman M.D., Collins T.J., Lipp P. Calcium signalling—an overview. Semin. Cell Dev. Biol. 2001;12:3–10. - PubMed
-
- Berridge M.J. Calcium signalling remodelling and disease. Biochem. Soc. Trans. 2012;40:297–309. - PubMed
-
- Berridge M.J., Bootman M.D., Lipp P. Calcium--a life and death signal. Nature. 1998;395:645–648. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

