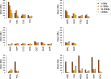Patterns of homozygosity in insular and continental goat breeds
- PMID: 30449277
- PMCID: PMC6241035
- DOI: 10.1186/s12711-018-0425-7
Patterns of homozygosity in insular and continental goat breeds
Abstract
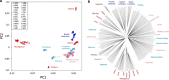
Background: Genetic isolation of breeds may result in a significant loss of diversity and have consequences on health and performance. In this study, we examined the effect of geographic isolation on caprine genetic diversity patterns by genotyping 480 individuals from 25 European and African breeds with the Goat SNP50 BeadChip and comparing patterns of homozygosity of insular and nearby continental breeds.
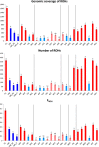
Results: Among the breeds analysed, number and total length of ROH varied considerably and depending on breeds, ROH could cover a substantial fraction of the genome (up to 1.6 Gb in Icelandic goats). When compared with their continental counterparts, goats from Iceland, Madagascar, La Palma and Ireland (Bilberry and Arran) displayed a significant increase in ROH coverage, ROH number and FROH values (P value < 0.05). Goats from Mediterranean islands represent a more complex case because certain populations displayed a significantly increased level of homozygosity (e.g. Girgentana) and others did not (e.g. Corse and Sarda). Correlations of number and total length of ROH for insular goat populations with the distance between islands and the nearest continental locations revealed an effect of extremely long distances on the patterns of homozygosity.
Conclusions: These results indicate that the effects of insularization on the patterns of homozygosity are variable. Goats raised in Madagascar, Iceland, Ireland (Bilberry and Arran) and La Palma, show high levels of homozygosity, whereas those bred in Mediterranean islands display patterns of homozygosity that are similar to those found in continental populations. These results indicate that the diversity of insular goat populations is modulated by multiple factors such as geographic distribution, population size, demographic history, trading and breed management.
Figures