Genome-wide SNP profiling of worldwide goat populations reveals strong partitioning of diversity and highlights post-domestication migration routes
- PMID: 30449284
- PMCID: PMC6240949
- DOI: 10.1186/s12711-018-0422-x
Genome-wide SNP profiling of worldwide goat populations reveals strong partitioning of diversity and highlights post-domestication migration routes
Abstract
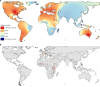
Background: Goat populations that are characterized within the AdaptMap project cover a large part of the worldwide distribution of this species and provide the opportunity to assess their diversity at a global scale. We analysed genome-wide 50 K single nucleotide polymorphism (SNP) data from 144 populations to describe the global patterns of molecular variation, compare them to those observed in other livestock species, and identify the drivers that led to the current distribution of goats.
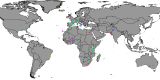
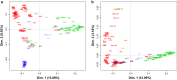
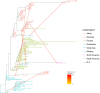
Results: A high degree of genetic variability exists among the goat populations studied. Our results highlight a strong partitioning of molecular diversity between and within continents. Three major gene pools correspond to goats from Europe, Africa and West Asia. Dissection of sub-structures disclosed regional gene pools, which reflect the main post-domestication migration routes. We also identified several exchanges, mainly in African populations, and which often involve admixed and cosmopolitan breeds. Extensive gene flow has taken place within specific areas (e.g., south Europe, Morocco and Mali-Burkina Faso-Nigeria), whereas elsewhere isolation due to geographical barriers (e.g., seas or mountains) or human management has decreased local gene flows.
Conclusions: After domestication in the Fertile Crescent in the early Neolithic era (ca. 12,000 YBP), domestic goats that already carried differentiated gene pools spread to Europe, Africa and Asia. The spread of these populations determined the major genomic background of the continental populations, which currently have a more marked subdivision than that observed in other ruminant livestock species. Subsequently, further diversification occurred at the regional level due to geographical and reproductive isolation, which was accompanied by additional migrations and/or importations, the traces of which are still detectable today. The effects of breed formation were clearly detected, particularly in Central and North Europe. Overall, our results highlight a remarkable diversity that occurs at the global scale and is locally partitioned and often affected by introgression from cosmopolitan breeds. These findings support the importance of long-term preservation of goat diversity, and provide a useful framework for investigating adaptive introgression, directing genetic improvement and choosing breeding targets.
Figures



References
-
- FAO. The second report on the state of the world’s animal genetic resources for food and agriculture. In: Scherf BD, Pilling D, editors. Rome: FAO Commission on Genetic Resources for Food and Agriculture Assessments; 2015. http://www.fao.org/3/a-i4787e/index.html. Accessed 15 August 2018.
-
- Gkiasta M, Russell T, Shennan S, Steele J. Neolithic transition in Europe: the radiocarbon record revisited. Antiquity. 2003;77:45–62. doi: 10.1017/S0003598X00061330. - DOI
-
- Hassan FA. Climate and cattle in North Africa: a first approximation. In: Blench RM, MacDonald KC, editors. The origins and development of African livestock: archaeology, genetics, linguistics and ethnography. London: UCL Press; 2000. pp. 61–86.

