Mitochondria-Derived Vesicles Deliver Antimicrobial Reactive Oxygen Species to Control Phagosome-Localized Staphylococcus aureus
- PMID: 30449314
- PMCID: PMC7323595
- DOI: 10.1016/j.chom.2018.10.005
Mitochondria-Derived Vesicles Deliver Antimicrobial Reactive Oxygen Species to Control Phagosome-Localized Staphylococcus aureus
Abstract
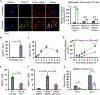
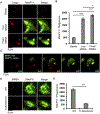
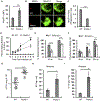
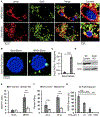
Pathogenic bacteria taken up into the macrophage phagosome are the target of many anti-microbial mechanisms. Although mitochondria-derived antimicrobial effectors like reactive oxygen species (mROS) aid in bacterial killing, it is unclear how these effectors reach bacteria within the phagosomal lumen. We show here that endoplasmic reticulum stress triggered upon methicillin-resistant Staphylococcus aureus (MRSA) infection induces mROS that are delivered to bacteria-containing phagosomes via mitochondria-derived vesicles (MDVs). The endoplasmic reticulum stress sensor IRE1α induces mROS, specifically hydrogen peroxide (mH2O2), upon MRSA infection. MRSA infection also stimulates the generation of MDVs, which require the mitochondrial stress response factor Parkin, and contributes to mH2O2 accumulation in bacteria-containing phagosomes. Accumulation of phagosomal H2O2 requires Toll-like receptor signaling and the mitochondrial enzyme superoxide dismutase-2 (Sod2), which is delivered to phagosomes by MDVs. Sod2 depletion compromises mH2O2 production and bacterial killing. Thus, mitochondrial redox capacity enhances macrophage antimicrobial function by delivering mitochondria-derived effector molecules into bacteria-containing phagosomes.
Copyright © 2018. Published by Elsevier Inc.
Conflict of interest statement
Declaration of Interests
The authors declare no competing interests.
Figures


Comment in
-
MDVs: Spare the SOD and Spoil the Bug.Cell Host Microbe. 2018 Nov 14;24(5):616-618. doi: 10.1016/j.chom.2018.10.013. Cell Host Microbe. 2018. PMID: 30439336
References
-
- Al-Qahtani AA, Al-Anazi MR, Al-Zoghaibi FA, Abdo AA, Sanai FM, Al-Hamoudi WK, Alswat KA, Al-Ashgar HI, Khan MQ, Albenmousa A, et al. (2016). PARK2 polymorphisms predict disease progression in patients infected with hepatitis C virus. Ann Hepatol 15, 824–833. - PubMed
-
- Bidani A, Reisner BS, Haque AK, Wen J, Helmer RE, Tuazon DM, and Heming TA (2000). Bactericidal activity of alveolar macrophages is suppressed by V-ATPase inhibition. Lung 178, 91–104. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

