Actionable Activating Oncogenic ERBB2/HER2 Transmembrane and Juxtamembrane Domain Mutations
- PMID: 30449325
- PMCID: PMC6248889
- DOI: 10.1016/j.ccell.2018.09.010
Actionable Activating Oncogenic ERBB2/HER2 Transmembrane and Juxtamembrane Domain Mutations
Abstract
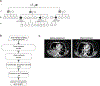
Deregulated HER2 is a target of many approved cancer drugs. We analyzed 111,176 patient tumors and identified recurrent mutations in HER2 transmembrane domain (TMD) and juxtamembrane domain (JMD) that include G660D, R678Q, E693K, and Q709L. Using a saturation mutagenesis screen and testing of patient-derived mutations we found several activating TMD and JMD mutations. Structural modeling and analysis showed that the TMD/JMD mutations function by improving the active dimer interface or stabilizing an activating conformation. Further, we found that HER2 G660D employed asymmetric kinase dimerization for activation and signaling. Importantly, anti-HER2 antibodies and small-molecule kinase inhibitors blocked the activity of TMD/JMD mutants. Consistent with this, a G660D germline mutant lung cancer patient showed remarkable clinical response to HER2 blockade.
Keywords: ERBB2 activation; ERBB2 structure; ERBB2/HER2; HER2 germline mutation; HER2 kinase inhibitors; HER2 somatic mutation; anti-HER2 antibodies; juxtamembrane (JMD) domain mutation; transmembrane domain (TMD) mutation.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures



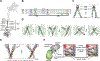
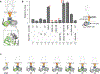

References
-
- Arteaga CL, Sliwkowski MX, Osborne CK, Perez EA, Puglisi F, and Gianni L (2011). Treatment of HER2-positive breast cancer: current status and future perspectives. Nat Rev Clin Oncol 9, 16–32. - PubMed
-
- Bargmann CI, Hung MC, and Weinberg RA (1986). Multiple independent activations of the neu oncogene by a point mutation altering the transmembrane domain of p185. Cell 45, 649657. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

