Genome-wide CRISPR Screens in Primary Human T Cells Reveal Key Regulators of Immune Function
- PMID: 30449619
- PMCID: PMC6689405
- DOI: 10.1016/j.cell.2018.10.024
Genome-wide CRISPR Screens in Primary Human T Cells Reveal Key Regulators of Immune Function
Abstract
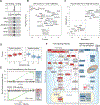
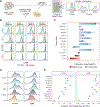
Human T cells are central effectors of immunity and cancer immunotherapy. CRISPR-based functional studies in T cells could prioritize novel targets for drug development and improve the design of genetically reprogrammed cell-based therapies. However, large-scale CRISPR screens have been challenging in primary human cells. We developed a new method, single guide RNA (sgRNA) lentiviral infection with Cas9 protein electroporation (SLICE), to identify regulators of stimulation responses in primary human T cells. Genome-wide loss-of-function screens identified essential T cell receptor signaling components and genes that negatively tune proliferation following stimulation. Targeted ablation of individual candidate genes characterized hits and identified perturbations that enhanced cancer cell killing. SLICE coupled with single-cell RNA sequencing (RNA-seq) revealed signature stimulation-response gene programs altered by key genetic perturbations. SLICE genome-wide screening was also adaptable to identify mediators of immunosuppression, revealing genes controlling responses to adenosine signaling. The SLICE platform enables unbiased discovery and characterization of functional gene targets in primary cells.
Keywords: CRISPR; T cell activation; T cell proliferation; genome-wide pooled screens; immunotherapy; primary human T cells; single-cell RNA-seq.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures



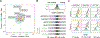
References
-
- Allard D, Turcotte M, and Stagg J (2017). Targeting A2 adenosine receptors in cancer. Immunol. Cell Biol. 95, 333–339. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

