Cryo-EM Structures of MDA5-dsRNA Filaments at Different Stages of ATP Hydrolysis
- PMID: 30449722
- PMCID: PMC6310684
- DOI: 10.1016/j.molcel.2018.10.012
Cryo-EM Structures of MDA5-dsRNA Filaments at Different Stages of ATP Hydrolysis
Abstract
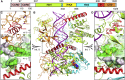
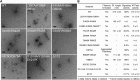
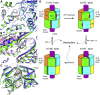
Double-stranded RNA (dsRNA) is a potent proinflammatory signature of viral infection. Long cytosolic dsRNA is recognized by MDA5. The cooperative assembly of MDA5 into helical filaments on dsRNA nucleates the assembly of a multiprotein type I interferon signaling platform. Here, we determined cryoelectron microscopy (cryo-EM) structures of MDA5-dsRNA filaments with different helical twists and bound nucleotide analogs at resolutions sufficient to build and refine atomic models. The structures identify the filament-forming interfaces, which encode the dsRNA binding cooperativity and length specificity of MDA5. The predominantly hydrophobic interface contacts confer flexibility, reflected in the variable helical twist within filaments. Mutation of filament-forming residues can result in loss or gain of signaling activity. Each MDA5 molecule spans 14 or 15 RNA base pairs, depending on the twist. Variations in twist also correlate with variations in the occupancy and type of nucleotide in the active site, providing insights on how ATP hydrolysis contributes to MDA5-dsRNA recognition.
Keywords: AGS; ATPase; Aicardi-Goutières syndrome; DExD/H-box RNA helicase; RIG-I-like receptor; RLR; SF2 helicases; SMS; Singleton-Merten syndrome; cryo-EM; cryoelectron microscopy; helical reconstruction; innate immune pattern recognition; nucleic acid sensing; superfamily 2 helicases.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Aricescu A.R., Lu W., Jones E.Y. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1243–1250. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

