Efficient method for isolation of reticulocyte RNA from healthy individuals and hemolytic anaemia patients
- PMID: 30450750
- PMCID: PMC6307756
- DOI: 10.1111/jcmm.13951
Efficient method for isolation of reticulocyte RNA from healthy individuals and hemolytic anaemia patients
Abstract
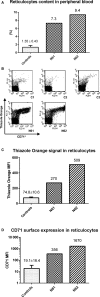
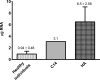
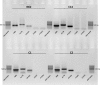
Despite enormous progress and development of high-throughput methods in genome-wide mRNA analyses, data on the erythroid transcriptome are still limited, even though they could be useful in medical diagnostics and personalized therapy as well as in research on normal and pathological erythroid maturation. Although obtaining normal and pathological reticulocyte transcriptome profiles should contribute greatly to our understanding of the molecular bases of terminal erythroid differentiation as well as the mechanisms of the hematological diseases, a basic limitation of these studies is the difficulty of efficient reticulocyte RNA isolation from human peripheral blood. The restricted number of possible parallel experiments primarily concern healthy individuals with the lowest number of reticulocytes in the peripheral blood and a low RNA content. In the present study, an efficient method for reticulocyte RNA isolation from healthy individuals and hemolytic anaemia patients is presented. The procedure includes leukofiltration, Ficoll-Paque gradient centrifugation, Percoll gradient centrifugation, and negative (CD45 and CD61) immunomagnetic separation. This relatively fast and simple four-stage method was successfully applied to obtain a reticulocyte-rich population from healthy subjects, which was used to efficiently isolate the high-quality RNA essential for successful NGS-based transcriptome analysis.
Keywords: RNA-Seq; hereditary hemolytic anaemia; hereditary spherocytosis; reticulocyte isolation; reticulocyte transcriptome.
© 2018 The Authors. Journal of Cellular and Molecular Medicine published by John Wiley & Sons Ltd and Foundation for Cellular and Molecular Medicine.
Figures


References
-
- Gronowicz G, Swift H, Steck TL. Maturation of the reticulocyte in vitro. J Cell Sci. 1984;71:177‐197. - PubMed
-
- Lee E, Choi HS, Hwang JH, Hoh JK, Cho YH, Baek EJ. The RNA in reticulocytes is not just debris: it is necessary for the final stages of erythrocyte formation. Blood Cells Mol Dis. 2014;53:1‐10. - PubMed
-
- d'Onofrio G, Kuse R, Foures C, Jou JM, Pradella M, Zini G. Reticulocytes in haematological disorders. Clin Lab Haematol. 1996;18(Suppl 1):29‐34. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

