Hidden diversity of soil giant viruses
- PMID: 30451857
- PMCID: PMC6243002
- DOI: 10.1038/s41467-018-07335-2
Hidden diversity of soil giant viruses
Abstract
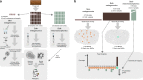
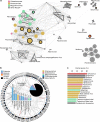
Known giant virus diversity is currently skewed towards viruses isolated from aquatic environments and cultivated in the laboratory. Here, we employ cultivation-independent metagenomics and mini-metagenomics on soils from the Harvard Forest, leading to the discovery of 16 novel giant viruses, chiefly recovered by mini-metagenomics. The candidate viruses greatly expand phylogenetic diversity of known giant viruses and either represented novel lineages or are affiliated with klosneuviruses, Cafeteria roenbergensis virus or tupanviruses. One assembled genome with a size of 2.4 Mb represents the largest currently known viral genome in the Mimiviridae, and others encode up to 80% orphan genes. In addition, we find more than 240 major capsid proteins encoded on unbinned metagenome fragments, further indicating that giant viruses are underexplored in soil ecosystems. The fact that most of these novel viruses evaded detection in bulk metagenomes suggests that mini-metagenomics could be a valuable approach to unearth viral giants.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

