Silibinin, A Natural Blend In Polytherapy Formulation For Targeting Cd44v6 Expressing Colon Cancer Stem Cells
- PMID: 30451890
- PMCID: PMC6242811
- DOI: 10.1038/s41598-018-35069-0
Silibinin, A Natural Blend In Polytherapy Formulation For Targeting Cd44v6 Expressing Colon Cancer Stem Cells
Erratum in
-
Publisher Correction: Silibinin, A Natural Blend In Polytherapy Formulation For Targeting Cd44v6 Expressing Colon Cancer Stem Cells.Sci Rep. 2018 Dec 17;8(1):17992. doi: 10.1038/s41598-018-36920-0. Sci Rep. 2018. PMID: 30559397 Free PMC article.
Abstract
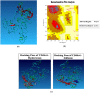
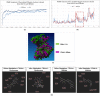
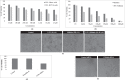
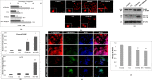
Colon cancer stem cells have been attributed to poor prognosis, therapeutic resistance and aggressive nature of the malignancy. Recent reports associated CD44v6 expression with relapse, metastasis and reduced 5-year survival of colon cancer patients, thereby making it a potential therapeutic target. Thus, in this study, comprehensive prediction and screening of CD44v6 against 1674 lead compounds was conducted. Silibinin was identified as a potential compound targeting CD44v6. Inorder to substantiate these findings, the cytotoxic effect of 5FU, Silibinin and 5FU+ Silibinin was assessed on human colon carcinoma cell line HCT116 derived CD44+ subpopulation. 5FU+ Silibinin inhibited cell proliferation of CD44+ subpopulation at lower concentration than Silibinin standalone. Further, corresponding to CD44v6 knockdown cells, 5FU+ Silibinin treatment significantly decreased CD44v6, Nanog, CTNNB1 and CDKN2A expression whereas increased E-cadherin expression in HCT116 derived CD44+ cells. Moreover, synergistic effect of these drugs suppressed sphere formation, inhibited cell migration, triggered PARP cleavage and perturbation in mitochondrial membrane potential, thereby activating intrinsic apoptotic pathways and induced autophagic cell death. Importantly, 5FU+ Silibinin could inhibit PI3K/MAPK dual activation and arrest the cell cycle at G0/G1 phase. Thus, our study suggests that inhibition of CD44v6 attenuates stemness of colon cancer stem cells and holds a prospect of potent therapeutic target.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Stewart, B.W. & Wild, C.P. editors. World Cancer Report 2014. Lyon, France: International Agency for Research on Cancer (2014). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

