RNA Recognition-like Motifs Activate a Mitogen-Activated Protein Kinase
- PMID: 30452242
- PMCID: PMC6457901
- DOI: 10.1021/acs.biochem.8b01032
RNA Recognition-like Motifs Activate a Mitogen-Activated Protein Kinase
Abstract
Smk1 is a mitogen-activated protein kinase (MAPK) family member in the yeast Saccharomyces cerevisiae that controls the postmeiotic program of spore formation. Ssp2 is a meiosis-specific protein that activates Smk1 and triggers the autophosphorylation of its activation loop. A fragment of Ssp2 that is sufficient to activate Smk1 contains two segments that resemble RNA recognition motifs (RRMs). Mutations in either of these motifs eliminated Ssp2's ability to activate Smk1. In contrast, deletions and insertions within the segment linking the RRM-like motifs only partially reduced the activity of Ssp2. Moreover, when the two RRM-like motifs were expressed as separate proteins in bacteria, they activated Smk1. We also find that both motifs can be cross-linked to Smk1 and that at least one of the motifs binds near the ATP-binding pocket of the MAPK. These findings demonstrate that motifs related to RRMs can directly activate protein kinases.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
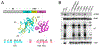



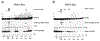
References
-
- Coulombe P, and Meloche S (2007) Atypical mitogen-activated protein kinases: structure, regulation and functions. Biochim. Biophys. Acta, Mol. Cell Res 1773, 1376–1387. - PubMed
-
- Pimienta G, and Pascual J (2007) Canonical and alternative MAPK signaling. Cell Cycle 6, 2628–2632. - PubMed
-
- Salvador JM, Mittelstadt PR, Guszczynski T, Copeland TD, Yamaguchi H, Appella E, Fornace AJ Jr., and Ashwell JD (2005) Alternative p38 activation pathway mediated by T cell receptor-proximal tyrosine kinases. Nat. Immunol 6, 390–395. - PubMed
-
- Ge B, Gram H, Di Padova F, Huang B, New L, Ulevitch RJ, Luo Y, and Han J (2002) MAPKK-independent activation of p38alpha mediated by TAB1-dependent autophosphorylation of p38alpha. Science 295, 1291–1294. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

