Metagenomics of pasteurized and unpasteurized gouda cheese using targeted 16S rDNA sequencing
- PMID: 30453904
- PMCID: PMC6245907
- DOI: 10.1186/s12866-018-1323-4
Metagenomics of pasteurized and unpasteurized gouda cheese using targeted 16S rDNA sequencing
Abstract
Background: The microbiome of cheese is diverse, even within a variety. The metagenomics of cheese is dependent on a vast array of biotic and abiotic factors. Biotic factors include the population of microbiota and their resulting cellular metabolism. Abiotic factors, including the pH, water activity, fat, salt, and moisture content of the cheese matrix, as well as environmental conditions (temperature, humidity, and location of aging), influence the biotic factors. This study assessed the metagenomics of commercial Gouda cheese prepared using pasteurized or unpasteurized cow milk or pasteurized goat milk via 16S rDNA sequencing.
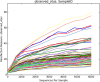
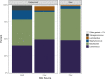
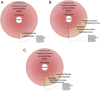
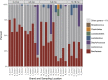
Results: Results were analyzed and compared based on milk pasteurization and source, spatial variability (core, outer, and under the rind), and length of aging (2-4 up to 12-18 months). The dominant organisms in the Gouda cheeses, based on percentage of sequence reads identified at the family or genus levels, were Bacillaceae, Lactococcus, Lactobacillus, Streptococcus, and Staphylococcus. More genus- or family-level (e.g. Bacillaceae) identifications were observed in the Gouda cheeses prepared with unpasteurized cow milk (120) compared with those prepared with pasteurized cow milk (92). When assessing influence of spatial variability on the metagenomics of the cheese, more pronounced differences in bacterial genera were observed in the samples taken under the rind; Brachybacterium, Pseudoalteromonas, Yersinia, Klebsiella, and Weissella were only detected in these samples. Lastly, the aging length of the cheese greatly influenced the number of organisms observed. Twenty-seven additional genus-level identifications were observed in Gouda cheese aged for 12-18 months compared with cheese only aged 2-4 months.
Conclusions: Collectively, the results of this study are important in determining the typical microbiota associated with Gouda cheese and how the microbiome plays a role in safety and quality.
Keywords: 16 s rDNA; Cheese; Dairy; Gouda; Metagenome; Unpasteurized milk.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Microbiota characterization of a Belgian protected designation of origin cheese, Herve cheese, using metagenomic analysis.J Dairy Sci. 2014 Oct;97(10):6046-56. doi: 10.3168/jds.2014-8225. Epub 2014 Jul 23. J Dairy Sci. 2014. PMID: 25064656
-
Microbial dynamics during the ripening of a mixed cow and goat milk cheese manufactured using frozen goat milk curd.J Dairy Sci. 2011 Oct;94(10):4766-76. doi: 10.3168/jds.2010-4077. J Dairy Sci. 2011. PMID: 21943728
-
Microbial community dynamics of a blue-veined raw milk cheese from the United Kingdom.J Dairy Sci. 2018 Jun;101(6):4923-4935. doi: 10.3168/jds.2017-14104. Epub 2018 Mar 15. J Dairy Sci. 2018. PMID: 29550118
-
Traditional cheeses: rich and diverse microbiota with associated benefits.Int J Food Microbiol. 2014 May 2;177:136-54. doi: 10.1016/j.ijfoodmicro.2014.02.019. Epub 2014 Mar 3. Int J Food Microbiol. 2014. PMID: 24642348 Review.
-
Milk microbiota: Characterization methods and role in cheese production.J Proteomics. 2020 Jan 6;210:103534. doi: 10.1016/j.jprot.2019.103534. Epub 2019 Oct 16. J Proteomics. 2020. PMID: 31629058 Review.
Cited by
-
Effect of reduction of sodium content on the microbial ecology of Edam cheese samples.AMB Express. 2021 Feb 16;11(1):28. doi: 10.1186/s13568-021-01188-7. AMB Express. 2021. PMID: 33591419 Free PMC article.
-
Evaluation of the kitchen microbiome and food safety behaviors of predominantly low-income families.Front Microbiol. 2022 Sep 29;13:987925. doi: 10.3389/fmicb.2022.987925. eCollection 2022. Front Microbiol. 2022. PMID: 36246211 Free PMC article.
-
Step-by-Step Metagenomics for Food Microbiome Analysis: A Detailed Review.Foods. 2024 Jul 14;13(14):2216. doi: 10.3390/foods13142216. Foods. 2024. PMID: 39063300 Free PMC article. Review.
-
High Throughput Sequencing Technologies as a New Toolbox for Deep Analysis, Characterization and Potentially Authentication of Protection Designation of Origin Cheeses?Int J Food Sci. 2019 Nov 20;2019:5837301. doi: 10.1155/2019/5837301. eCollection 2019. Int J Food Sci. 2019. PMID: 31886165 Free PMC article. Review.
-
The Invisible Threat of Antibiotic Resistance in Food.Antibiotics (Basel). 2025 Mar 1;14(3):250. doi: 10.3390/antibiotics14030250. Antibiotics (Basel). 2025. PMID: 40149061 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

