Prediction of Subgenome Additive and Interaction Effects in Allohexaploid Wheat
- PMID: 30455185
- PMCID: PMC6404612
- DOI: 10.1534/g3.118.200613
Prediction of Subgenome Additive and Interaction Effects in Allohexaploid Wheat
Abstract
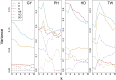
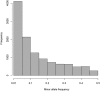
Whole genome duplications have played an important role in the evolution of angiosperms. These events often occur through hybridization between closely related species, resulting in an allopolyploid with multiple subgenomes. With the availability of affordable genotyping and a reference genome to locate markers, breeders of allopolyploids now have the opportunity to manipulate subgenomes independently. This also presents a unique opportunity to investigate epistatic interactions between homeologous orthologs across subgenomes. We present a statistical framework for partitioning genetic variance to the subgenomes of an allopolyploid, predicting breeding values for each subgenome, and determining the importance of inter-genomic epistasis. We demonstrate using an allohexaploid wheat breeding population evaluated in Ithaca, NY and an important wheat dataset from CIMMYT previously shown to demonstrate non-additive genetic variance. Subgenome covariance matrices were constructed and used to calculate subgenome interaction covariance matrices for variance component estimation and genomic prediction. We propose a method to extract population structure from all subgenomes at once before covariances are calculated to reduce collinearity between subgenome estimates. Variance parameter estimation was shown to be reliable for additive subgenome effects, but was less reliable for subgenome interaction components. Predictive ability was equivalent to current genomic prediction methods. Including only inter-genomic interactions resulted in the same increase in accuracy as modeling all pairwise marker interactions. Thus, we provide a new tool for breeders of allopolyploid crops to characterize the genetic architecture of existing populations, determine breeding goals, and develop new strategies for selection of additive effects and fixation of inter-genomic epistasis.
Keywords: Allopolyploidy; Epistasis; Genomic prediction; Heterosis.
Copyright © 2019 Santantonio et al.
Figures



References
-
- Abel S., Möllers C., Becker H., 2005. Development of synthetic brassica napus lines for the analysis of “fixed heterosis” in allopolyploid plants. Euphytica 146: 157–163. 10.1007/s10681-005-3364-7 - DOI
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
