Diagnostics of short tandem repeat expansion variants using massively parallel sequencing and componential tools
- PMID: 30455479
- PMCID: PMC6460572
- DOI: 10.1038/s41431-018-0302-4
Diagnostics of short tandem repeat expansion variants using massively parallel sequencing and componential tools
Abstract
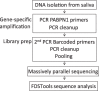
Short tandem repeats (STRs) are scattered throughout the human genome. Some STRs, like trinucleotide repeat expansion (TRE) variants, cause hereditable disorders. Unambiguous molecular diagnostics of TRE disorders is hampered by current technical limitations imposed by traditional PCR and DNA sequencing methods. Here we report a novel pipeline for TRE variant diagnosis employing the massively parallel sequencing (MPS) combined with an opensource software package (FDSTools), which together are designed to distinguish true STR sequences from STR sequencing artifacts. We show that this approach can improve TRE diagnosis, such as Oculopharyngeal muscular dystrophy (OPMD). OPMD is caused by a trinucleotide expansion in the PABPN1 gene. A short GCN expansion, (GCN[10]), coding for a 10 alanine repeat is not pathogenic, but an alanine expansion is pathogenic. Applying this novel procedure in a Dutch OPMD patient cohort, we found expansion variants from GCN[11] to GCN[16], with the GCN[16] as the most abundant variant. The repeat expansion length did not correlate with clinical features. However, symptom severity was found to correlate with age and with the initial affected muscles, suggesting that aging and muscle-specific factors can play a role in modulating OPMD.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

