Human Dendritic Cell Subsets Undergo Distinct Metabolic Reprogramming for Immune Response
- PMID: 30455688
- PMCID: PMC6230993
- DOI: 10.3389/fimmu.2018.02489
Human Dendritic Cell Subsets Undergo Distinct Metabolic Reprogramming for Immune Response
Abstract
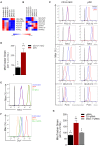
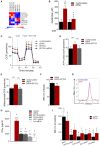
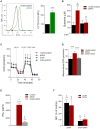
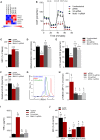
Toll-like receptor (TLR) agonists induce metabolic reprogramming, which is required for immune activation. We have investigated mechanisms that regulate metabolic adaptation upon TLR-stimulation in human blood DC subsets, CD1c+ myeloid DCs (mDCs) and plasmacytoid DCs (pDCs). We show that TLR-stimulation changes expression of genes regulating oxidative phosphorylation (OXPHOS) and glutamine metabolism in pDC. TLR-stimulation increases mitochondrial content and intracellular glutamine in an autophagy-dependent manner in pDC. TLR-induced glutaminolysis fuels OXPHOS in pDCs. Notably, inhibition of glutaminolysis and OXPHOS prevents pDC activation. Conversely, TLR-stimulation reduces mitochondrial content, OXPHOS activity and induces glycolysis in CD1c+ mDC. Inhibition of mitochondrial fragmentation or promotion of mitochondrial fusion impairs TLR-stimulation induced glycolysis and activation of CD1c+ mDCs. TLR-stimulation triggers BNIP3-dependent mitophagy, which regulates transcriptional activity of AMPKα1. BNIP3-dependent mitophagy is required for induction of glycolysis and activation of CD1c+ mDCs. Our findings reveal that TLR stimulation differentially regulates mitochondrial dynamics in distinct human DC subsets, which contributes to their activation.
Keywords: CD1c+ mDC; OXPHOS; glutaminolysis; glycolysis; mitochondrial dynamics; mitophagy; pDC.
Figures

Similar articles
-
Human Plasmacytoid and Monocyte-Derived Dendritic Cells Display Distinct Metabolic Profile Upon RIG-I Activation.Front Immunol. 2018 Dec 21;9:3070. doi: 10.3389/fimmu.2018.03070. eCollection 2018. Front Immunol. 2018. PMID: 30622542 Free PMC article.
-
A Comparative Study of the T Cell Stimulatory and Polarizing Capacity of Human Primary Blood Dendritic Cell Subsets.Mediators Inflamm. 2016;2016:3605643. doi: 10.1155/2016/3605643. Epub 2016 Feb 7. Mediators Inflamm. 2016. PMID: 27057096 Free PMC article.
-
Dendritic Cells Require PINK1-Mediated Phosphorylation of BCKDE1α to Promote Fatty Acid Oxidation for Immune Function.Front Immunol. 2019 Oct 15;10:2386. doi: 10.3389/fimmu.2019.02386. eCollection 2019. Front Immunol. 2019. PMID: 31681280 Free PMC article.
-
The role of nitric oxide in metabolic regulation of Dendritic cell immune function.Cancer Lett. 2018 Jan 1;412:236-242. doi: 10.1016/j.canlet.2017.10.032. Epub 2017 Oct 26. Cancer Lett. 2018. PMID: 29107106 Free PMC article. Review.
-
Metabolic Reprogramming in Mitochondria of Myeloid Cells.Cells. 2019 Dec 18;9(1):5. doi: 10.3390/cells9010005. Cells. 2019. PMID: 31861356 Free PMC article. Review.
Cited by
-
Immuno-metabolic dendritic cell vaccine signatures associate with overall survival in vaccinated melanoma patients.Nat Commun. 2023 Nov 8;14(1):7211. doi: 10.1038/s41467-023-42881-4. Nat Commun. 2023. PMID: 37938561 Free PMC article.
-
Immunometabolism in atherosclerotic disorders.Nat Cardiovasc Res. 2024 Jun;3(6):637-650. doi: 10.1038/s44161-024-00473-5. Epub 2024 May 23. Nat Cardiovasc Res. 2024. PMID: 39196223 Review.
-
The circadian clock influences T cell responses to vaccination by regulating dendritic cell antigen processing.Nat Commun. 2022 Dec 5;13(1):7217. doi: 10.1038/s41467-022-34897-z. Nat Commun. 2022. PMID: 36470865 Free PMC article.
-
The role of dendritic cells and their immunometabolism in rheumatoid arthritis.Front Immunol. 2023 May 12;14:1161148. doi: 10.3389/fimmu.2023.1161148. eCollection 2023. Front Immunol. 2023. PMID: 37251399 Free PMC article. Review.
-
Endosomal trafficking inhibitor EGA can control TLR7-mediated IFNα expression by human plasmacytoid dendritic cells.Front Immunol. 2023 Nov 24;14:1202197. doi: 10.3389/fimmu.2023.1202197. eCollection 2023. Front Immunol. 2023. PMID: 38077311 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

