Haplotype Loci Under Selection in Canadian Durum Wheat Germplasm Over 60 Years of Breeding: Association With Grain Yield, Quality Traits, Protein Loss, and Plant Height
- PMID: 30455711
- PMCID: PMC6230583
- DOI: 10.3389/fpls.2018.01589
Haplotype Loci Under Selection in Canadian Durum Wheat Germplasm Over 60 Years of Breeding: Association With Grain Yield, Quality Traits, Protein Loss, and Plant Height
Abstract
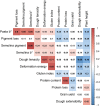
Durum wheat was introduced in the southern prairies of western Canada in the late nineteenth century. Breeding efforts have mainly focused on improving quality traits to meet the pasta industry demands. For this study, 192 durum wheat lines were genotyped using the Illumina 90K Infinium iSelect assay, and resulted in a total of 14,324 polymorphic SNPs. Genetic diversity changed over time, declining during the first 20 years of breeding in Canada, then increased in the late 1980s and early 1990s. We scanned the genome for signatures of selection, using the total variance Fst-based outlier detection method (Lositan), the hierarchical island model (Arlequin) and the Bayesian genome scan method (BayeScan). A total of 407 outliers were identified and clustered into 84 LD-based haplotype loci, spanning all 14 chromosomes of the durum wheat genome. The association analysis detected 54 haplotype loci, of which 39% contained markers with a complete reversal of allelic state. This tendency to fixation of favorable alleles corroborates the success of the Canadian durum wheat breeding programs over time. Twenty-one haplotype loci were associated with multiple traits. In particular, hap_4B_1 explained 20.6, 17.9 and 16.6% of the phenotypic variance of pigment loss, pasta b∗ and dough extensibility, respectively. The locus hap_2B_9 explained 15.9 and 17.8% of the variation of protein content and protein loss, respectively. All these pleiotropic haplotype loci offer breeders the unique opportunity for further improving multiple traits, facilitating marker-assisted selection in durum wheat, and could help in identifying genes as functional annotations of the wheat genome become available.
Keywords: durum wheat; grain yield; haplotype; loci under selection; plant height; protein loss; quality traits.
Figures




Similar articles
-
Carotenoid Pigment Content in Durum Wheat (Triticum turgidum L. var durum): An Overview of Quantitative Trait Loci and Candidate Genes.Front Plant Sci. 2019 Nov 7;10:1347. doi: 10.3389/fpls.2019.01347. eCollection 2019. Front Plant Sci. 2019. PMID: 31787991 Free PMC article. Review.
-
Single Marker and Haplotype-Based Association Analysis of Semolina and Pasta Colour in Elite Durum Wheat Breeding Lines Using a High-Density Consensus Map.PLoS One. 2017 Jan 30;12(1):e0170941. doi: 10.1371/journal.pone.0170941. eCollection 2017. PLoS One. 2017. PMID: 28135299 Free PMC article.
-
Whole Genome Scan Reveals Molecular Signatures of Divergence and Selection Related to Important Traits in Durum Wheat Germplasm.Front Genet. 2020 Apr 21;11:217. doi: 10.3389/fgene.2020.00217. eCollection 2020. Front Genet. 2020. PMID: 32373150 Free PMC article.
-
Association Mapping for 24 Traits Related to Protein Content, Gluten Strength, Color, Cooking, and Milling Quality Using Balanced and Unbalanced Data in Durum Wheat [Triticum turgidum L. var. durum (Desf).].Front Genet. 2019 Aug 16;10:717. doi: 10.3389/fgene.2019.00717. eCollection 2019. Front Genet. 2019. PMID: 31475032 Free PMC article.
-
From ancient to old and modern durum wheat varieties: interaction among cultivar traits, management, and technological quality.J Sci Food Agric. 2019 Mar 30;99(5):2059-2067. doi: 10.1002/jsfa.9388. Epub 2018 Nov 8. J Sci Food Agric. 2019. PMID: 30267406 Review.
Cited by
-
From Genetic Maps to QTL Cloning: An Overview for Durum Wheat.Plants (Basel). 2021 Feb 6;10(2):315. doi: 10.3390/plants10020315. Plants (Basel). 2021. PMID: 33562160 Free PMC article. Review.
-
Carotenoid Pigment Content in Durum Wheat (Triticum turgidum L. var durum): An Overview of Quantitative Trait Loci and Candidate Genes.Front Plant Sci. 2019 Nov 7;10:1347. doi: 10.3389/fpls.2019.01347. eCollection 2019. Front Plant Sci. 2019. PMID: 31787991 Free PMC article. Review.
-
Genomic prediction of agronomic traits in wheat using different models and cross-validation designs.Theor Appl Genet. 2021 Jan;134(1):381-398. doi: 10.1007/s00122-020-03703-z. Epub 2020 Nov 1. Theor Appl Genet. 2021. PMID: 33135095
-
Genomics for Yield and Yield Components in Durum Wheat.Plants (Basel). 2023 Jul 7;12(13):2571. doi: 10.3390/plants12132571. Plants (Basel). 2023. PMID: 37447132 Free PMC article. Review.
-
Meta-analysis: Congruence of genomic and phenotypic differentiation across diverse natural study systems.Evol Appl. 2021 Aug 19;14(9):2189-2205. doi: 10.1111/eva.13264. eCollection 2021 Sep. Evol Appl. 2021. PMID: 34603492 Free PMC article.
References
-
- Acreche M. M., Slafer G. A. (2009). Variation of grain nitrogen content in relation with grain yield in old and modern Spanish wheats grown under a wide range of agronomic conditions in a Mediterranean region. J. Agric. Sci. 147 657–667. 10.1017/S0021859609990190 - DOI
-
- Aremu C. O. (2011). Genetic diversity: a review for need and measurements for intraspecies crop improvement. J. Microbiol. Biotech. Res. 1 80–85.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

