Chronic platelet-derived growth factor receptor signaling exerts control over initiation of protein translation in glioma
- PMID: 30456354
- PMCID: PMC6238596
- DOI: 10.26508/lsa.201800029
Chronic platelet-derived growth factor receptor signaling exerts control over initiation of protein translation in glioma
Abstract
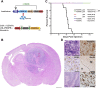
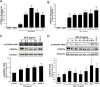
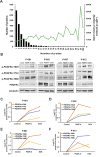
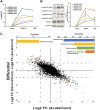
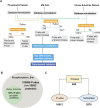
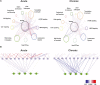
Activation of the platelet-derived growth factor receptors (PDGFRs) gives rise to some of the most important signaling pathways that regulate mammalian cellular growth, survival, proliferation, and differentiation and their misregulation is common in a variety of diseases. Herein, we present a comprehensive and detailed map of PDGFR signaling pathways assembled from literature and integrate this map in a bioinformatics protocol designed to extract meaningful information from large-scale quantitative proteomics mass spectrometry data. We demonstrate the usefulness of this approach using a new genetically engineered mouse model of PDGFRα-driven glioma. We discovered that acute PDGFRα stimulation differs considerably from chronic receptor activation in the regulation of protein translation initiation. Transient stimulation activates several key components of the translation initiation machinery, whereas the clinically relevant chronic activity of PDGFRα is associated with a significant shutdown of translational members. Our work defines a step-by-step approach to extract biologically relevant insights from global unbiased phospho-protein datasets to uncover targets for therapeutic assessment.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures








References
-
- Acquaviva J, Jun HJ, Lessard J, Ruiz R, Zhu H, Donovan M, Woolfenden S,Boskovitz A, Raval A, Bronson RT, et al. (2011) Chronic activation of wild-type epidermal growth factor receptor and loss of Cdkn2a cause mouse glioblastoma formation. Cancer Res 71: 7198–7206. 10.1158/0008-5472.can-11-1514 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
