A stochastic epigenetic switch controls the dynamics of T-cell lineage commitment
- PMID: 30457103
- PMCID: PMC6245732
- DOI: 10.7554/eLife.37851
A stochastic epigenetic switch controls the dynamics of T-cell lineage commitment
Abstract
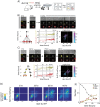
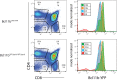
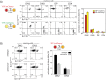
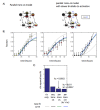
Cell fate decisions occur through the switch-like, irreversible activation of fate-specifying genes. These activation events are often assumed to be tightly coupled to changes in upstream transcription factors, but could also be constrained by cis-epigenetic mechanisms at individual gene loci. Here, we studied the activation of Bcl11b, which controls T-cell fate commitment. To disentangle cis and trans effects, we generated mice where two Bcl11b copies are tagged with distinguishable fluorescent proteins. Quantitative live microscopy of progenitors from these mice revealed that Bcl11b turned on after a stochastic delay averaging multiple days, which varied not only between cells but also between Bcl11b alleles within the same cell. Genetic perturbations, together with mathematical modeling, showed that a distal enhancer controls the rate of epigenetic activation, while a parallel Notch-dependent trans-acting step stimulates expression from activated loci. These results show that developmental fate transitions can be controlled by stochastic cis-acting events on individual loci.
Keywords: computational biology; developmental biology; epigenetics; gene regulation; lymphocyte development; mouse; quantitative and systems biology; stochastic gene expression; systems biology.
© 2018, Ng et al.
Conflict of interest statement
KN, MY, AM, SS, BI, SP, SH, ME, ER, HK No competing interests declared
Figures













Similar articles
-
A timed epigenetic switch balances T and ILC lineage proportions in the thymus.Development. 2024 Dec 1;151(23):dev203016. doi: 10.1242/dev.203016. Epub 2024 Dec 10. Development. 2024. PMID: 39655434 Free PMC article.
-
Asynchronous combinatorial action of four regulatory factors activates Bcl11b for T cell commitment.Nat Immunol. 2016 Aug;17(8):956-65. doi: 10.1038/ni.3514. Epub 2016 Jul 4. Nat Immunol. 2016. PMID: 27376470 Free PMC article.
-
Bcl11b and combinatorial resolution of cell fate in the T-cell gene regulatory network.Proc Natl Acad Sci U S A. 2017 Jun 6;114(23):5800-5807. doi: 10.1073/pnas.1610617114. Proc Natl Acad Sci U S A. 2017. PMID: 28584128 Free PMC article.
-
Epigenetic Dynamics in the Function of T-Lineage Regulatory Factor Bcl11b.Front Immunol. 2021 Apr 14;12:669498. doi: 10.3389/fimmu.2021.669498. eCollection 2021. Front Immunol. 2021. PMID: 33936112 Free PMC article. Review.
-
T Cell Development by the Numbers.Trends Immunol. 2017 Feb;38(2):128-139. doi: 10.1016/j.it.2016.10.007. Epub 2016 Nov 12. Trends Immunol. 2017. PMID: 27842955 Review.
Cited by
-
Stage-specific action of Runx1 and GATA3 controls silencing of PU.1 expression in mouse pro-T cells.J Exp Med. 2021 Aug 2;218(8):e20202648. doi: 10.1084/jem.20202648. Epub 2021 Jun 28. J Exp Med. 2021. PMID: 34180951 Free PMC article.
-
Causal Gene Regulatory Network Modeling and Genomics: Second-Generation Challenges.J Comput Biol. 2019 Jul;26(7):703-718. doi: 10.1089/cmb.2019.0098. Epub 2019 May 7. J Comput Biol. 2019. PMID: 31063008 Free PMC article.
-
Semicoordinated allelic-bursting shape dynamic random monoallelic expression in pregastrulation embryos.iScience. 2021 Aug 5;24(9):102954. doi: 10.1016/j.isci.2021.102954. eCollection 2021 Sep 24. iScience. 2021. PMID: 34458702 Free PMC article.
-
Using computational modelling to reveal mechanisms of epigenetic Polycomb control.Biochem Soc Trans. 2021 Feb 26;49(1):71-77. doi: 10.1042/BST20190955. Biochem Soc Trans. 2021. PMID: 33616630 Free PMC article. Review.
-
Enhancer Priming Enables Fast and Sustained Transcriptional Responses to Notch Signaling.Dev Cell. 2019 Aug 19;50(4):411-425.e8. doi: 10.1016/j.devcel.2019.07.002. Epub 2019 Aug 1. Dev Cell. 2019. PMID: 31378591 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

