The lncRNA RMEL3 protects immortalized cells from serum withdrawal-induced growth arrest and promotes melanoma cell proliferation and tumor growth
- PMID: 30457212
- PMCID: PMC6613776
- DOI: 10.1111/pcmr.12751
The lncRNA RMEL3 protects immortalized cells from serum withdrawal-induced growth arrest and promotes melanoma cell proliferation and tumor growth
Erratum in
-
Corrigendum.Pigment Cell Melanoma Res. 2020 May;33(3):520. doi: 10.1111/pcmr.12875. Pigment Cell Melanoma Res. 2020. PMID: 32319742 No abstract available.
Abstract
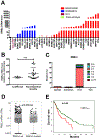
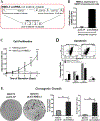
RMEL3 is a recently identified lncRNA associated with BRAFV600E mutation and melanoma cell survival. Here, we demonstrate strong and moderate RMEL3 upregulation in BRAF and NRAS mutant melanoma cells, respectively, compared to melanocytes. High expression is also more frequent in cutaneous than in acral/mucosal melanomas, and analysis of an ICGC melanoma dataset showed that mutations in RMEL3 locus are preponderantly C > T substitutions at dipyrimidine sites including CC > TT, typical of UV signature. RMEL3 mutation does not correlate with RMEL3 levels, but does with poor patient survival, in TCGA melanoma dataset. Accordingly, RMEL3 lncRNA levels were significantly reduced in BRAFV600E melanoma cells upon treatment with BRAF or MEK inhibitors, supporting the notion that BRAF-MEK-ERK pathway plays a role to activate RMEL3 gene transcription. RMEL3 overexpression, in immortalized fibroblasts and melanoma cells, increased proliferation and survival under serum starvation, clonogenic ability, and xenografted melanoma tumor growth. Although future studies will be needed to elucidate the mechanistic activities of RMEL3, our data demonstrate that its overexpression bypasses the need of mitogen activation to sustain proliferation/survival of non-transformed cells and suggest an oncogenic role for RMEL3.
Keywords: BRAFV600E; CTD-2023N9.1; ENSG00000250961.1; LncGPBP1-1:1; MAPK; chr5:57395060-57533424 (GRCh38/hg38); melanoma; mitogen; serum response.
© 2018 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- 457603/2013-5/Conselho Nacional de Desenvolvimento Científico e Tecnológico/International
- 465687/2014-8/Conselho Nacional de Desenvolvimento Científico e Tecnológico/International
- 870310/1997-6/Conselho Nacional de Desenvolvimento Científico e Tecnológico/International
- R01 AR072304/AR/NIAMS NIH HHS/United States
- 2014/18189-5/Fundação de Amparo à Pesquisa do Estado de São Paulo/International
- 309187/2015-0/Conselho Nacional de Desenvolvimento Científico e Tecnológico/International
- P01 CA163222/CA/NCI NIH HHS/United States
- 2012/24056-2/Fundação de Amparo à Pesquisa do Estado de São Paulo/International
- 2010/16097-5/Fundação de Amparo à Pesquisa do Estado de São Paulo/International
- R01 CA222871/CA/NCI NIH HHS/United States
- R01 AR043369/AR/NIAMS NIH HHS/United States
- 2013/08135-2/Fundação de Amparo à Pesquisa do Estado de São Paulo/International
- 2018/04017-9/Fundação de Amparo à Pesquisa do Estado de São Paulo/International
- 2014/50928-2/Fundação de Amparo à Pesquisa do Estado de São Paulo/International
- 2013/13465-1/Fundação de Amparo à Pesquisa do Estado de São Paulo/International
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

