A Cell/Cilia Cycle Biosensor for Single-Cell Kinetics Reveals Persistence of Cilia after G1/S Transition Is a General Property in Cells and Mice
- PMID: 30458140
- PMCID: PMC6251972
- DOI: 10.1016/j.devcel.2018.10.027
A Cell/Cilia Cycle Biosensor for Single-Cell Kinetics Reveals Persistence of Cilia after G1/S Transition Is a General Property in Cells and Mice
Abstract
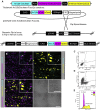
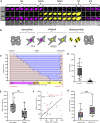
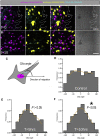
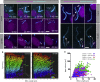
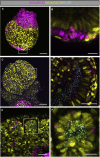
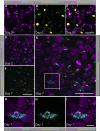
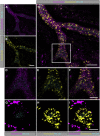
The cilia and cell cycles are inextricably linked. Centrioles in the basal body of cilia nucleate the ciliary axoneme and sequester pericentriolar matrix (PCM) at the centrosome to organize the mitotic spindle. Cilia themselves respond to growth signals, prompting cilia resorption and cell cycle re-entry. We describe a fluorescent cilia and cell cycle biosensor allowing live imaging of cell cycle progression and cilia assembly and disassembly kinetics in cells and inducible mice. We define assembly and disassembly in relation to cell cycle stage with single-cell resolution and explore the intercellular heterogeneity in cilia kinetics. In all cells and tissues analyzed, we observed cilia that persist through the G1/S transition and into S/G2/M-phase. We conclude that persistence of cilia after the G1/S transition is a general property. This resource will shed light at an individual cell level on the interplay between the cilia and cell cycles in development, regeneration, and disease.
Keywords: Rosa26; biosensor; cell cycle; cilia; live imaging; organoid; reporter mouse.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Live Imaging and Analysis of Cilia and Cell Cycle Dynamics with the Arl13bCerulean-Fucci2a Biosensor and Fucci Tools.Methods Mol Biol. 2021;2329:291-309. doi: 10.1007/978-1-0716-1538-6_21. Methods Mol Biol. 2021. PMID: 34085231
-
PCM1 recruits Plk1 to the pericentriolar matrix to promote primary cilia disassembly before mitotic entry.J Cell Sci. 2013 Mar 15;126(Pt 6):1355-65. doi: 10.1242/jcs.114918. Epub 2013 Jan 23. J Cell Sci. 2013. PMID: 23345402
-
CEP162 is an axoneme-recognition protein promoting ciliary transition zone assembly at the cilia base.Nat Cell Biol. 2013 Jun;15(6):591-601. doi: 10.1038/ncb2739. Epub 2013 May 5. Nat Cell Biol. 2013. PMID: 23644468 Free PMC article.
-
Regulating the transition from centriole to basal body.J Cell Biol. 2011 May 2;193(3):435-44. doi: 10.1083/jcb.201101005. J Cell Biol. 2011. PMID: 21536747 Free PMC article. Review.
-
The Centrioles, Centrosomes, Basal Bodies, and Cilia of Drosophila melanogaster.Genetics. 2017 May;206(1):33-53. doi: 10.1534/genetics.116.198168. Genetics. 2017. PMID: 28476861 Free PMC article. Review.
Cited by
-
Dynamic Changes in Ultrastructure of the Primary Cilium in Migrating Neuroblasts in the Postnatal Brain.J Neurosci. 2019 Dec 11;39(50):9967-9988. doi: 10.1523/JNEUROSCI.1503-19.2019. Epub 2019 Nov 4. J Neurosci. 2019. PMID: 31685650 Free PMC article.
-
Insights into the Regulation of Ciliary Disassembly.Cells. 2021 Nov 1;10(11):2977. doi: 10.3390/cells10112977. Cells. 2021. PMID: 34831200 Free PMC article. Review.
-
TTBK2 controls cilium stability by regulating distinct modules of centrosomal proteins.Mol Biol Cell. 2023 Jan 1;34(1):ar8. doi: 10.1091/mbc.E22-08-0373. Epub 2022 Nov 2. Mol Biol Cell. 2023. PMID: 36322399 Free PMC article.
-
DeCOOC Deconvoluted Hi-C Map Characterizes the Chromatin Architecture of Cells in Physiologically Distinctive Tissues.Adv Sci (Weinh). 2023 Sep;10(27):e2301058. doi: 10.1002/advs.202301058. Epub 2023 Jul 28. Adv Sci (Weinh). 2023. PMID: 37515382 Free PMC article.
-
Structure, function, and research progress of primary cilia in reproductive physiology and reproductive diseases.Front Cell Dev Biol. 2024 Jun 3;12:1418928. doi: 10.3389/fcell.2024.1418928. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38887518 Free PMC article. Review.
References
-
- Abe T., Sakaue-Sawano A., Kiyonari H., Shioi G., Inoue K., Horiuchi T., Nakao K., Miyawaki A., Aizawa S., Fujimori T. Visualization of cell cycle in mouse embryos with Fucci2 reporter directed by Rosa26 promoter. Development. 2013;140:237–246. - PubMed
-
- Bellusci S., Furuta Y., Rush M.G., Henderson R., Winnier G., Hogan B.L. Involvement of Sonic hedgehog (Shh) in mouse embryonic lung growth and morphogenesis. Development. 1997;124:53–63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MC_PC_U127574433/MRC_/Medical Research Council/United Kingdom
- MC_PC_U127527200/MRC_/Medical Research Council/United Kingdom
- NC/M001091/1/MRC_/Medical Research Council/United Kingdom
- NC/M001091/1/NC3RS_/National Centre for the Replacement, Refinement and Reduction of Animals in Research/United Kingdom
- MC_UU_00007/17/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

