Metatranscriptomes Reveal That All Three Domains of Life Are Active but Are Dominated by Bacteria in the Fennoscandian Crystalline Granitic Continental Deep Biosphere
- PMID: 30459191
- PMCID: PMC6247080
- DOI: 10.1128/mBio.01792-18
Metatranscriptomes Reveal That All Three Domains of Life Are Active but Are Dominated by Bacteria in the Fennoscandian Crystalline Granitic Continental Deep Biosphere
Abstract
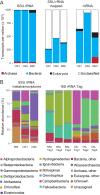
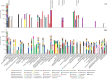
The continental subsurface is suggested to contain a significant part of the earth's total biomass. However, due to the difficulty of sampling, the deep subsurface is still one of the least understood ecosystems. Therefore, microorganisms inhabiting this environment might profoundly influence the global nutrient and energy cycles. In this study, in situ fixed RNA transcripts from two deep continental groundwaters from the Äspö Hard Rock Laboratory (a Baltic Sea-influenced water with a residence time of <20 years, defined as "modern marine," and an "old saline" groundwater with a residence time of thousands of years) were subjected to metatranscriptome sequencing. Although small subunit (SSU) rRNA gene and mRNA transcripts aligned to all three domains of life, supporting activity within these community subsets, the data also suggested that the groundwaters were dominated by bacteria. Many of the SSU rRNA transcripts grouped within newly described candidate phyla or could not be mapped to known branches on the tree of life, suggesting that a large portion of the active biota in the deep biosphere remains unexplored. Despite the extremely oligotrophic conditions, mRNA transcripts revealed a diverse range of metabolic strategies that were carried out by multiple taxa in the modern marine water that is fed by organic carbon from the surface. In contrast, the carbon dioxide- and hydrogen-fed old saline water with a residence time of thousands of years predominantly showed the potential to carry out translation. This suggested these cells were active, but waiting until an energy source episodically becomes available.IMPORTANCE A newly designed sampling apparatus was used to fix RNA under in situ conditions in the deep continental biosphere and benchmarks a strategy for deep biosphere metatranscriptomic sequencing. This apparatus enabled the identification of active community members and the processes they carry out in this extremely oligotrophic environment. This work presents for the first time evidence of eukaryotic, archaeal, and bacterial activity in two deep subsurface crystalline rock groundwaters from the Äspö Hard Rock Laboratory with different depths and geochemical characteristics. The findings highlight differences between organic carbon-fed shallow communities and carbon dioxide- and hydrogen-fed old saline waters. In addition, the data reveal a large portion of uncharacterized microorganisms, as well as the important role of candidate phyla in the deep biosphere, but also the disparity in microbial diversity when using standard microbial 16S rRNA gene amplification versus the large unknown portion of the community identified with unbiased metatranscriptomes.
Keywords: deep biosphere; groundwaters; mRNA; metatranscriptomes; rRNA.
Copyright © 2018 Lopez-Fernandez et al.
Figures


Similar articles
-
Statistical Analysis of Community RNA Transcripts between Organic Carbon and Geogas-Fed Continental Deep Biosphere Groundwaters.mBio. 2019 Aug 13;10(4):e01470-19. doi: 10.1128/mBio.01470-19. mBio. 2019. PMID: 31409677 Free PMC article.
-
Depth and Dissolved Organic Carbon Shape Microbial Communities in Surface Influenced but Not Ancient Saline Terrestrial Aquifers.Front Microbiol. 2018 Nov 27;9:2880. doi: 10.3389/fmicb.2018.02880. eCollection 2018. Front Microbiol. 2018. PMID: 30538690 Free PMC article.
-
Investigation of viable taxa in the deep terrestrial biosphere suggests high rates of nutrient recycling.FEMS Microbiol Ecol. 2018 Aug 1;94(8):fiy121. doi: 10.1093/femsec/fiy121. FEMS Microbiol Ecol. 2018. PMID: 29931252 Free PMC article.
-
The deep continental subsurface: the dark biosphere.Int Microbiol. 2018 Jun;21(1-2):3-14. doi: 10.1007/s10123-018-0009-y. Epub 2018 May 30. Int Microbiol. 2018. PMID: 30810923 Review.
-
Prokaryotic biodiversity and activity in the deep subseafloor biosphere.FEMS Microbiol Ecol. 2008 Nov;66(2):181-96. doi: 10.1111/j.1574-6941.2008.00566.x. Epub 2008 Aug 20. FEMS Microbiol Ecol. 2008. PMID: 18752622 Review.
Cited by
-
Biofilm Formation on Excavation Damaged Zone Fractures in Deep Neogene Sedimentary Rock.Microb Ecol. 2024 Oct 22;87(1):132. doi: 10.1007/s00248-024-02451-7. Microb Ecol. 2024. PMID: 39436423 Free PMC article.
-
Active anaerobic methane oxidation and sulfur disproportionation in the deep terrestrial subsurface.ISME J. 2022 Jun;16(6):1583-1593. doi: 10.1038/s41396-022-01207-w. Epub 2022 Feb 16. ISME J. 2022. PMID: 35173296 Free PMC article.
-
Dark biosphere: Just at the very tip of the iceberg.Environ Microbiol. 2023 Jan;25(1):147-149. doi: 10.1111/1462-2920.16265. Epub 2022 Nov 9. Environ Microbiol. 2023. PMID: 36307896 Free PMC article. No abstract available.
-
Novel candidate taxa contribute to key metabolic processes in Fennoscandian Shield deep groundwaters.ISME Commun. 2024 Sep 23;4(1):ycae113. doi: 10.1093/ismeco/ycae113. eCollection 2024 Jan. ISME Commun. 2024. PMID: 39421601 Free PMC article.
-
Optimization of Mapping Tools and Investigation of Ribosomal RNA Influence for Data-Driven Gene Expression Analysis in Complex Microbiomes.Microorganisms. 2025 Apr 26;13(5):995. doi: 10.3390/microorganisms13050995. Microorganisms. 2025. PMID: 40431168 Free PMC article.
References
-
- Heuer VB, Pohlman JW, Torres ME, Elvert M, Hinrichs K-U. 2009. The stable carbon isotope biogeochemistry of acetate and other dissolved carbon species in deep subseafloor sediments at the northern Cascadia Margin. Geochim Cosmochim Acta 73:3323–3336. doi:10.1016/j.gca.2009.03.001. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
