Cryptococcus neoformans Cda1 and Its Chitin Deacetylase Activity Are Required for Fungal Pathogenesis
- PMID: 30459196
- PMCID: PMC6247093
- DOI: 10.1128/mBio.02087-18
Cryptococcus neoformans Cda1 and Its Chitin Deacetylase Activity Are Required for Fungal Pathogenesis
Abstract
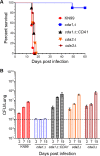
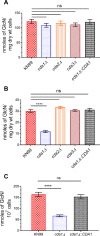
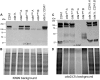
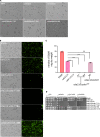
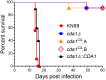
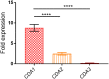
Chitin is an essential component of the cell wall of Cryptococcus neoformans conferring structural rigidity and integrity under diverse environmental conditions. Chitin deacetylase genes encode the enyzmes (chitin deacetylases [Cdas]) that deacetylate chitin, converting it to chitosan. The functional role of chitosan in the fungal cell wall is not well defined, but it is an important virulence determinant of C. neoformans Mutant strains deficient in chitosan are completely avirulent in a mouse pulmonary infection model. C. neoformans carries genes that encode three Cdas (Cda1, Cda2, and Cda3) that appear to be functionally redundant in cells grown under vegetative conditions. Here we report that C. neoformans Cda1 is the principal Cda responsible for fungal pathogenesis. Point mutations were introduced in the active site of Cda1 to generate strains in which the enzyme activity of Cda1 was abolished without perturbing either its stability or localization. When used to infect CBA/J mice, Cda1 mutant strains produced less chitosan and were attenuated for virulence. We further demonstrate that C. neoformans Cda genes are transcribed differently during a murine infection from what has been measured in vitroIMPORTANCECryptococcus neoformans is unique among fungal pathogens that cause disease in a mammalian host, as it secretes a polysaccharide capsule that hinders recognition by the host to facilitate its survival and proliferation. Even though it causes serious infections in immunocompromised hosts, reports of infection in hosts that are immunocompetent are on the rise. The cell wall of a fungal pathogen, its synthesis, composition, and pathways of remodelling are attractive therapeutic targets for the development of fungicides. Chitosan, a polysaccharide in the cell wall of C. neoformans is one such target, as it is critical for pathogenesis and absent in the host. The results we present shed light on the importance of one of the chitin deacetylases that synthesize chitosan during infection and further implicates chitosan as being a critical factor for the pathogenesis of C. neoformans.
Keywords: catalytic site; cell wall localization; chitin deacetylase; chitosan; fungal virulence; metal binding site.
Copyright © 2018 Upadhya et al.
Figures



Similar articles
-
Chitosan Biosynthesis and Virulence in the Human Fungal Pathogen Cryptococcus gattii.mSphere. 2019 Oct 9;4(5):e00644-19. doi: 10.1128/mSphere.00644-19. mSphere. 2019. PMID: 31597720 Free PMC article.
-
Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans.Eukaryot Cell. 2007 May;6(5):855-67. doi: 10.1128/EC.00399-06. Epub 2007 Mar 30. Eukaryot Cell. 2007. PMID: 17400891 Free PMC article.
-
Evolutionary analysis and protein family classification of chitin deacetylases in Cryptococcus neoformans.J Microbiol. 2020 Sep;58(9):805-811. doi: 10.1007/s12275-020-0288-9. Epub 2020 Sep 1. J Microbiol. 2020. PMID: 32870486
-
Connecting virulence pathways to cell-cycle progression in the fungal pathogen Cryptococcus neoformans.Curr Genet. 2017 Oct;63(5):803-811. doi: 10.1007/s00294-017-0688-5. Epub 2017 Mar 6. Curr Genet. 2017. PMID: 28265742 Free PMC article. Review.
-
Cryptococcus neoformans: a sugar-coated killer with designer genes.FEMS Immunol Med Microbiol. 2005 Sep 1;45(3):395-404. doi: 10.1016/j.femsim.2005.06.005. FEMS Immunol Med Microbiol. 2005. PMID: 16055314 Review.
Cited by
-
Aspartyl peptidase May1 induces host inflammatory response by altering cell wall composition in the fungal pathogen Cryptococcus neoformans.mBio. 2024 Jun 12;15(6):e0092024. doi: 10.1128/mbio.00920-24. Epub 2024 May 14. mBio. 2024. PMID: 38742885 Free PMC article.
-
The Aminoalkylindole BML-190 Negatively Regulates Chitosan Synthesis via the Cyclic AMP/Protein Kinase A1 Pathway in Cryptococcus neoformans.mBio. 2019 Dec 17;10(6):e02264-19. doi: 10.1128/mBio.02264-19. mBio. 2019. PMID: 31848271 Free PMC article.
-
Powerful cell wall biomass degradation enzymatic system from saprotrophic Aspergillus fumigatus.Cell Surf. 2024 May 21;11:100126. doi: 10.1016/j.tcsw.2024.100126. eCollection 2024 Jun. Cell Surf. 2024. PMID: 38827922 Free PMC article. Review.
-
Measuring Stress Phenotypes in Cryptococcus neoformans.Methods Mol Biol. 2024;2775:277-303. doi: 10.1007/978-1-0716-3722-7_19. Methods Mol Biol. 2024. PMID: 38758325 Free PMC article.
-
Harnessing the Immune Response to Fungal Pathogens for Vaccine Development.Annu Rev Microbiol. 2022 Sep 8;76:703-726. doi: 10.1146/annurev-micro-041020-111511. Epub 2022 Jun 27. Annu Rev Microbiol. 2022. PMID: 35759871 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

