Human Stress-inducible Hsp70 Has a High Propensity to Form ATP-dependent Antiparallel Dimers That Are Differentially Regulated by Cochaperone Binding
- PMID: 30459217
- PMCID: PMC6356074
- DOI: 10.1074/mcp.RA118.001044
Human Stress-inducible Hsp70 Has a High Propensity to Form ATP-dependent Antiparallel Dimers That Are Differentially Regulated by Cochaperone Binding
Abstract
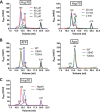
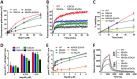
Eukaryotic protein homeostasis (proteostasis) is largely dependent on the action of highly conserved Hsp70 molecular chaperones. Recent evidence indicates that, apart from conserved molecular allostery, Hsp70 proteins have retained and adapted the ability to assemble as functionally relevant ATP-bound dimers throughout evolution. Here, we have compared the ATP-dependent dimerization of DnaK, human stress-inducible Hsp70, Hsc70 and BiP Hsp70 proteins, showing that their dimerization propensities differ, with stress-inducible Hsp70 being predominantly dimeric in the presence of ATP. Structural analyses using hydrogen/deuterium exchange mass spectrometry, native electrospray ionization mass spectrometry and small-angle X-ray scattering revealed that stress-inducible Hsp70 assembles in solution as an antiparallel dimer with the intermolecular interface closely resembling the ATP-bound dimer interfaces captured in DnaK and BiP crystal structures. ATP-dependent dimerization of stress-inducible Hsp70 is necessary for its efficient interaction with Hsp40, as shown by experiments with dimerization-deficient mutants. Moreover, dimerization of ATP-bound Hsp70 is required for its participation in high molecular weight protein complexes detected ex vivo, supporting its functional role in vivo As human cytosolic Hsp70 can interact with tetratricopeptide repeat (TPR) domain containing cochaperones, we tested the interaction of Hsp70 ATP-dependent dimers with Chip and Tomm34 cochaperones. Although Chip associates with intact Hsp70 dimers to form a larger complex, binding of Tomm34 disrupts the Hsp70 dimer and this event plays an important role in Hsp70 activity regulation. In summary, this study provides structural evidence of robust ATP-dependent antiparallel dimerization of human inducible Hsp70 protein and suggests a novel role of TPR domain cochaperones in multichaperone complexes involving Hsp70 ATP-bound dimers.
Keywords: Allostery; Chaperone; Cochaperone; Mass Spectrometry; Protein Conformation; Protein Structure; Protein-Protein Interactions; Structural Biology.
© 2019 Trcka et al.
Figures







References
-
- Young J. C., Agashe V. R., Siegers K., and Hartl F. U. (2004) Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 5, 781–791 - PubMed
-
- Boorstein W. R., Ziegelhoffer T., and Craig E. A. (1994) Molecular evolution of the HSP70 multigene family. J. Mol. Evolution 38, 1–17 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

