Interactome mapping defines BRG1, a component of the SWI/SNF chromatin remodeling complex, as a new partner of the transcriptional regulator CTCF
- PMID: 30459231
- PMCID: PMC6341399
- DOI: 10.1074/jbc.RA118.004882
Interactome mapping defines BRG1, a component of the SWI/SNF chromatin remodeling complex, as a new partner of the transcriptional regulator CTCF
Abstract
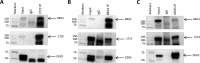
The highly conserved zinc finger CCCTC-binding factor (CTCF) regulates genomic imprinting and gene expression by acting as a transcriptional activator or repressor of promoters and insulator of enhancers. The multiple functions of CTCF are accomplished by co-association with other protein partners and are dependent on genomic context and tissue specificity. Despite the critical role of CTCF in the organization of genome structure, to date, only a subset of CTCF interaction partners have been identified. Here we present a large-scale identification of CTCF-binding partners using affinity purification and high-resolution LC-MS/MS analysis. In addition to functional enrichment of specific protein families such as the ribosomal proteins and the DEAD box helicases, we identified novel high-confidence CTCF interactors that provide a still unexplored biochemical context for CTCF's multiple functions. One of the newly validated CTCF interactors is BRG1, the major ATPase subunit of the chromatin remodeling complex SWI/SNF, establishing a relationship between two master regulators of genome organization. This work significantly expands the current knowledge of the human CTCF interactome and represents an important resource to direct future studies aimed at uncovering molecular mechanisms modulating CTCF pleiotropic functions throughout the genome.
Keywords: BRG1; CTCF; Interactomics; SWI/SNF; chromatin; mass spectrometry (MS); protein–protein interaction; proteomics; transcription factor; transcriptional regulation.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




Similar articles
-
Exploring the Interaction between the SWI/SNF Chromatin Remodeling Complex and the Zinc Finger Factor CTCF.Int J Mol Sci. 2020 Nov 25;21(23):8950. doi: 10.3390/ijms21238950. Int J Mol Sci. 2020. PMID: 33255744 Free PMC article.
-
Novel Interactions between the Human T-Cell Leukemia Virus Type 1 Antisense Protein HBZ and the SWI/SNF Chromatin Remodeling Family: Implications for Viral Life Cycle.J Virol. 2019 Jul 30;93(16):e00412-19. doi: 10.1128/JVI.00412-19. Print 2019 Aug 15. J Virol. 2019. PMID: 31142665 Free PMC article.
-
Cdx2 Regulates Gene Expression through Recruitment of Brg1-associated Switch-Sucrose Non-fermentable (SWI-SNF) Chromatin Remodeling Activity.J Biol Chem. 2017 Feb 24;292(8):3389-3399. doi: 10.1074/jbc.M116.752774. Epub 2017 Jan 12. J Biol Chem. 2017. PMID: 28082674 Free PMC article.
-
The BRG1 ATPase of human SWI/SNF chromatin remodeling enzymes as a driver of cancer.Epigenomics. 2017 Jun;9(6):919-931. doi: 10.2217/epi-2017-0034. Epub 2017 May 19. Epigenomics. 2017. PMID: 28521512 Free PMC article. Review.
-
Involvement of the chromatin-remodeling factor BRG1/SMARCA4 in human cancer.Epigenetics. 2008 Mar-Apr;3(2):64-8. doi: 10.4161/epi.3.2.6153. Epub 2008 Apr 17. Epigenetics. 2008. PMID: 18437052 Review.
Cited by
-
Exploring the Interaction between the SWI/SNF Chromatin Remodeling Complex and the Zinc Finger Factor CTCF.Int J Mol Sci. 2020 Nov 25;21(23):8950. doi: 10.3390/ijms21238950. Int J Mol Sci. 2020. PMID: 33255744 Free PMC article.
-
Cancer-specific CTCF binding facilitates oncogenic transcriptional dysregulation.Genome Biol. 2020 Sep 15;21(1):247. doi: 10.1186/s13059-020-02152-7. Genome Biol. 2020. PMID: 32933554 Free PMC article.
-
Mithramycin induces promoter reprogramming and differentiation of rhabdoid tumor.EMBO Mol Med. 2021 Feb 5;13(2):e12640. doi: 10.15252/emmm.202012640. Epub 2020 Dec 17. EMBO Mol Med. 2021. PMID: 33332735 Free PMC article.
-
CTCF and Its Multi-Partner Network for Chromatin Regulation.Cells. 2023 May 10;12(10):1357. doi: 10.3390/cells12101357. Cells. 2023. PMID: 37408191 Free PMC article. Review.
-
At the Crossroad of Gene Regulation and Genome Organization: Potential Roles for ATP-Dependent Chromatin Remodelers in the Regulation of CTCF-Mediated 3D Architecture.Biology (Basel). 2021 Mar 27;10(4):272. doi: 10.3390/biology10040272. Biology (Basel). 2021. PMID: 33801596 Free PMC article. Review.
References
-
- Kanduri C., Pant V., Loukinov D., Pugacheva E., Qi C. F., Wolffe A., Ohlsson R., and Lobanenkov V. V. (2000) Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 10, 853–856 10.1074/jbc.RA117.001068 - DOI - PubMed
-
- Filippova G. N., Fagerlie S., Klenova E. M., Myers C., Dehner Y., Goodwin G., Neiman P. E., Collins S. J., and Lobanenkov V. V. (1996) An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol. 16, 2802–2813 10.1074/jbc.RA117.001068 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

