The ubiquitin ligase SspH1 from Salmonella uses a modular and dynamic E3 domain to catalyze substrate ubiquitylation
- PMID: 30459234
- PMCID: PMC6341402
- DOI: 10.1074/jbc.RA118.004247
The ubiquitin ligase SspH1 from Salmonella uses a modular and dynamic E3 domain to catalyze substrate ubiquitylation
Abstract
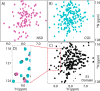
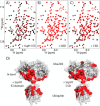
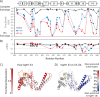
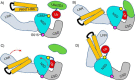
SspH/IpaH bacterial effector E3 ubiquitin (Ub) ligases, unrelated in sequence or structure to eukaryotic E3s, are utilized by a wide variety of Gram-negative bacteria during pathogenesis. These E3s function in a eukaryotic environment, utilize host cell E2 ubiquitin-conjugating enzymes of the Ube2D family, and target host proteins for ubiquitylation. Despite several crystal structures, details of Ube2D∼Ub binding and the mechanism of ubiquitin transfer are poorly understood. Here, we show that the catalytic E3 ligase domain of SspH1 can be divided into two subdomains: an N-terminal subdomain that harbors the active-site cysteine and a C-terminal subdomain containing the Ube2D∼Ub-binding site. SspH1 mutations designed to restrict subdomain motions show rapid formation of an E3∼Ub intermediate, but impaired Ub transfer to substrate. NMR experiments using paramagnetic spin labels reveal how SspH1 binds Ube2D∼Ub and targets the E2∼Ub active site. Unexpectedly, hydrogen/deuterium exchange MS shows that the E2∼Ub-binding region is dynamic but stabilized in the E3∼Ub intermediate. Our results support a model in which both subunits of an Ube2D∼Ub clamp onto a dynamic region of SspH1, promoting an E3 conformation poised for transthiolation. A conformational change is then required for Ub transfer from E3∼Ub to substrate.
Keywords: PKN1; Salmonella; SspH1; Ube2D; bacterial effector; bacterial pathogenesis; ubiquitin; ubiquitin ligase (E3 enzyme); ubiquitin transfer; ubiquitin-conjugating enzyme (E2 enzyme); ubiquitylation (ubiquitination).
© 2019 Cook et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

