The inflammasome adapter ASC assembles into filaments with integral participation of its two Death Domains, PYD and CARD
- PMID: 30459235
- PMCID: PMC6333874
- DOI: 10.1074/jbc.RA118.004407
The inflammasome adapter ASC assembles into filaments with integral participation of its two Death Domains, PYD and CARD
Abstract
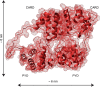
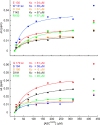
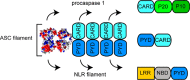
The inflammasome is a multiprotein complex necessary for the onset of inflammation. The adapter protein ASC assembles inflammasome components by acting as a molecular glue between danger-signal sensors and procaspase-1. The assembly is mediated by ASC self-association and protein interactions via its two Death Domains, PYD and CARD. Truncated versions of ASC have been shown to form filaments, but information on the filaments formed by full-length ASC is needed to construct a meaningful model of inflammasome assembly. To gain insights into this system, we used a combination of transmission EM, NMR, and computational analysis to investigate intact ASC structures. We show that ASC forms ∼6-7-nm-wide filaments that stack laterally to form bundles. The structural characteristics and dimensions of the bundles indicate that both PYD and CARD are integral parts of the filament. A truncated version of ASC with only the CARD domain (ASCCARD) forms different filaments (∼3-4-nm width), providing further evidence that both domains work in concert in filament assembly. Ring-shaped protein particles bound to pre-existing filaments match the size of ASC dimer structures generated by NMR-based protein docking, suggesting that the ASC dimer could be a basic building block for filament formation. Solution NMR binding studies identified the protein surfaces involved in the ASCCARD-ASCCARD interaction. These data provide new insights into the structural underpinnings of the inflammasome and should inform future efforts to interrogate this important biological system.
Keywords: ASC; CARD; Death Domain; NMR; PYD; apoptosis; inflammasome; inflammation; protein assembly; transmission electron microscopy.
© 2019 Nambayan et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures










References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

