Precise tracking of vaccine-responding T cell clones reveals convergent and personalized response in identical twins
- PMID: 30459272
- PMCID: PMC6294963
- DOI: 10.1073/pnas.1809642115
Precise tracking of vaccine-responding T cell clones reveals convergent and personalized response in identical twins
Abstract
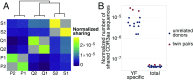
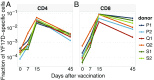
T cell receptor (TCR) repertoire data contain information about infections that could be used in disease diagnostics and vaccine development, but extracting that information remains a major challenge. Here we developed a statistical framework to detect TCR clone proliferation and contraction from longitudinal repertoire data. We applied this framework to data from three pairs of identical twins immunized with the yellow fever vaccine. We identified 600 to 1,700 responding TCRs in each donor and validated them using three independent assays. While the responding TCRs were mostly private, albeit with higher overlap between twins, they could be well-predicted using a classifier based on sequence similarity. Our method can also be applied to samples obtained postinfection, making it suitable for systematic discovery of new infection-specific TCRs in the clinic.
Keywords: RepSeq; T cell receptor; high-throughput sequencing; twins; vaccination.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

