Tissue-Based Mapping of the Fathead Minnow (Pimephales promelas) Transcriptome and Proteome
- PMID: 30459712
- PMCID: PMC6232228
- DOI: 10.3389/fendo.2018.00611
Tissue-Based Mapping of the Fathead Minnow (Pimephales promelas) Transcriptome and Proteome
Abstract
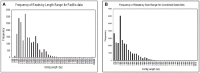
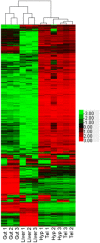
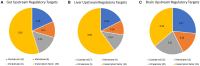
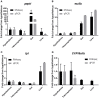
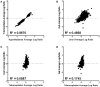
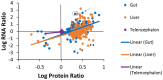
Omics approaches are broadly used to explore endocrine and toxicity-related pathways and functions. Nevertheless, there is still a significant gap in knowledge in terms of understanding the endocrine system and its numerous connections and intricate feedback loops, especially in non-model organisms. The fathead minnow (Pimephales promelas) is a widely used small fish model for aquatic toxicology and regulatory testing, particularly in North America. A draft genome has been published, but the amount of available genomic or transcriptomic information is still far behind that of other more broadly studied species, such as the zebrafish. Here, we used a proteogenomics approach to survey the tissue-specific proteome and transcriptome profiles in adult male fathead minnow. To do so, we generated a draft transcriptome using short and long sequencing reads from liver, testis, brain, heart, gill, head kidney, trunk kidney, and gastrointestinal tract. We identified 30,378 different putative transcripts overall, with the assembled contigs ranging in size from 264 to over 9,720 nts. Over 17,000 transcripts were >1,000 nts, suggesting a robust transcriptome that can be used to interpret RNA sequencing data in the future. We also performed RNA sequencing and proteomics analysis on four tissues, including the telencephalon, hypothalamus, liver, and gastrointestinal tract of male fish. Transcripts ranged from 0 to 600,000 copies per gene and a large portion were expressed in a tissue-specific manner. Specifically, the telencephalon and hypothalamus shared the most expressed genes, while the gastrointestinal tract and the liver were quite distinct. Using protein profiling techniques, we identified a total of 4,045 proteins in the four tissues investigated, and their tissue-specific expression pattern correlated with the transcripts at the pathway level. Similarly to the findings with the transcriptomic data, the hypothalamus and telencephalon had the highest degree of similarity in the proteins detected. The main purpose of this analysis was to generate tissue-specific omics data in order to support future aquatic ecotoxicogenomic and endocrine-related studies as well as to improve our understanding of the fathead minnow as an ecological model.
Keywords: endocrine system; fathead minnow; proteogenomics; proteome; tissue-specific; transcriptome.
Figures

Similar articles
-
A graphical systems model and tissue-specific functional gene sets to aid transcriptomic analysis of chemical impacts on the female teleost reproductive axis.Mutat Res. 2012 Aug 15;746(2):151-62. doi: 10.1016/j.mrgentox.2011.12.016. Epub 2011 Dec 28. Mutat Res. 2012. PMID: 22227403
-
Advancing the fathead minnow (Pimephales promelas) as a model for immunotoxicity testing: Characterization of the renal transcriptome following Yersinia ruckeri infection.Fish Shellfish Immunol. 2020 Aug;103:472-480. doi: 10.1016/j.fsi.2020.05.008. Epub 2020 May 18. Fish Shellfish Immunol. 2020. PMID: 32439514
-
Natural Variation in Fish Transcriptomes: Comparative Analysis of the Fathead Minnow (Pimephales promelas) and Zebrafish (Danio rerio).PLoS One. 2014 Dec 10;9(12):e114178. doi: 10.1371/journal.pone.0114178. eCollection 2014. PLoS One. 2014. PMID: 25493933 Free PMC article.
-
The fathead minnow in aquatic toxicology: past, present and future.Aquat Toxicol. 2006 Jun 10;78(1):91-102. doi: 10.1016/j.aquatox.2006.01.018. Epub 2006 Feb 21. Aquat Toxicol. 2006. PMID: 16494955 Review.
-
Small fish models for identifying and assessing the effects of endocrine-disrupting chemicals.ILAR J. 2004;45(4):469-83. doi: 10.1093/ilar.45.4.469. ILAR J. 2004. PMID: 15454686 Review.
Cited by
-
Comparative Transcriptomic and Expression Profiles Between the Foot Muscle and Mantle Tissues in the Giant Triton Snail Charonia tritonis.Front Physiol. 2021 Feb 24;12:632518. doi: 10.3389/fphys.2021.632518. eCollection 2021. Front Physiol. 2021. PMID: 33732164 Free PMC article.
-
De Novo Assembly of the Nearly Complete Fathead Minnow Reference Genome Reveals a Repetitive but Compact Genome.Environ Toxicol Chem. 2022 Feb;41(2):448-461. doi: 10.1002/etc.5266. Epub 2022 Jan 18. Environ Toxicol Chem. 2022. PMID: 34888930 Free PMC article.
-
Cross-Taxa Distinctions in Mechanisms of Developmental Effects for Aquatic Species Exposed to Trifluralin.Environ Toxicol Chem. 2020 Sep;39(9):1797-1812. doi: 10.1002/etc.4758. Epub 2020 Jul 7. Environ Toxicol Chem. 2020. PMID: 32445211 Free PMC article.
-
Transcriptomics and proteomics provide insights into the adaptative strategies of Tibetan naked carps (Gymnocypris przewalskii) to saline-alkaline variations.BMC Genomics. 2025 Feb 19;26(1):162. doi: 10.1186/s12864-025-11336-z. BMC Genomics. 2025. PMID: 39972273 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

