Novel Self-Transmissible and Broad-Host-Range Plasmids Exogenously Captured From Anaerobic Granules or Cow Manure
- PMID: 30459733
- PMCID: PMC6232296
- DOI: 10.3389/fmicb.2018.02602
Novel Self-Transmissible and Broad-Host-Range Plasmids Exogenously Captured From Anaerobic Granules or Cow Manure
Abstract
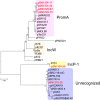
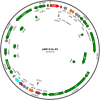
Novel self-transmissible plasmids were exogenously captured from environmental samples by triparental matings with pBBR1MCS-2 as a mobilizable plasmid and Pseudomonas resinovorans as a recipient. A total of 272 recipients were successfully obtained as plasmid host candidates from granules of an anaerobic methane fermentation plant and from cow manure. The whole nucleotide sequences of six plasmids were determined, including one IncP-1 plasmid (pSN1104-59), four PromA-like plasmids (pSN1104-11, pSN1104-34, pSN0729-62, and pSN0729-70), and one novel plasmid (pSN1216-29), whose incompatibility group has not been previously identified. No previously known antibiotic resistance genes were found in these plasmids. In-depth phylogenetic analyses showed that the PromA-like plasmids belong to subgroups of PromA (designated as PromAγ and PromAδ) different from previously proposed subgroups PromAα and PromAβ. Twenty-four genes were identified as backbone genes by comparisons with other PromA plasmids. The nucleotide sequences of pSN1216-29 share high identity with those found in clinical isolates. A minireplicon of pSN1216-29 was successfully constructed from repA encoding a replication initiation protein and oriV. All the captured plasmids were found to have a broad host range and could be transferred to and replicated in different classes of Proteobacteria. Notably, repA and oriV of pSN1216-29 showed high similarity with one of two replication systems of pSRC119-A/C, known as a plasmid with multidrug resistance genes found in Salmonella enterica serovar Senftenberg. Our findings suggest that these "cryptic" but broad-host-range plasmids may be important for spreading several genes as "vehicles" in a wider range of bacteria in natural environments.
Keywords: PromA; broad host range; conjugation; plasmid; replication.
Figures



References
-
- Arakawa K., Suzuki H., Tomita M. (2008). Computational genome analysis using the G-language system. Genes Genomes Genomics 2 1–13.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

