Identification of a Novel QTL for Panicle Length From Wild Rice (Oryza minuta) by Specific Locus Amplified Fragment Sequencing and High Density Genetic Mapping
- PMID: 30459776
- PMCID: PMC6232755
- DOI: 10.3389/fpls.2018.01492
Identification of a Novel QTL for Panicle Length From Wild Rice (Oryza minuta) by Specific Locus Amplified Fragment Sequencing and High Density Genetic Mapping
Abstract
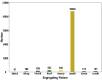
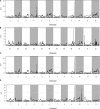
Wild rice possesses a large number of valuable genes that have been lost or do not exist in cultivated rice. To exploit the desirable gene controlling panicle length (PL) in wild rice Oryza minuta, a recombinant inbred line (RIL) population was constructed that was derived from a cross between the long panicle introgression line K1561 with Oryza minuta segments and a short panicle accession G1025. Specific Locus Amplified Fragment (SLAF) sequencing technology was used to uncover single nucleotide polymorphisms (SNPs) and construct the high-density genetic linkage map. Using 201 RIL populations, a high-density genetic map was developed, and spanned 2781.76 cM with an average genetic distance 0.45 cM. The genetic map was composed of 5, 521 markers on 12 chromosomes. Based on this high-density genome map, quantitative trait loci (QTL) for PL were analyzed for 2 years under four environments. Seven QTLs were detected, which were distributed within chromosomes 4, 9, and 10, respectively. pl4.1 was detected twice, and pl10.1 was only detected once. Although pl9.1 was only detected once, it was very near pl9.2 in the genetic map which was detected three times. Thus, we speculate one major QTL exists in the region of pl9.1 and pl9.2 to control PL (temporarily referred to as pl9). pl9 is a potentially novel allele derived from Oryza minuta, and it can be used for genetic improvement of cultivar rice.
Keywords: RILs; SNPs; introgression lines; linkage map; yield related traits.
Figures



References
-
- Guo S. (2009). Development and Characterization of Substation Lines from a Cross of Oryza sativa and Oryza minuta. Dissertation thesis, Huzhong Agricultural University, Wuhan.
LinkOut - more resources
Full Text Sources

