Autophagy mediates SUMO-induced degradation of a polyglutamine protein ataxin-3
- PMID: 30460066
- PMCID: PMC6138331
- DOI: 10.1080/19768354.2017.1330765
Autophagy mediates SUMO-induced degradation of a polyglutamine protein ataxin-3
Abstract
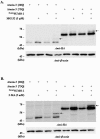
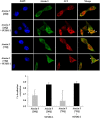
Previously, we reported that small ubiquitin-like modifier-1 (SUMO-1) promotes the degradation of a polyglutamine (polyQ) protein ataxin-3 and proposed that proteasomes mediate the proteolysis. Here, we present evidence that autophagy is also responsible for SUMO-induced degradation of this polyQ protein. The autophagy inhibitor 3-MA increased the steady-state level of ataxin-3 and stabilized SUMO-modified ataxin-3 more prominently than the proteasome inhibitor MG132. Interestingly, SUMO-1 overexpression enhanced the co-localization of ataxin-3 and autophagy marker LC3 without increasing LC3 puncta formation suggesting that SUMO-1 is involved in the substrate recruitment rather than the induction of autophagy. To assess the importance of a putative SUMO-interacting motif (SIM) in ataxin-3 for SUMO-induced degradation, we constructed a SIM mutant of ataxin-3. Substitution of putative SIM (V165G) facilitated the degradation of polyQ-expanded ataxin-3, which is more resistant to SUMO-induced degradation than the normal ataxin-3. These results together indicate that SUMO-1 promotes the degradation of ataxin-3 via autophagy and the putative SIM of ataxin-3 plays a role in this process.
Keywords: Ataxin-3; SUMO; SUMO-interacting motif; autophagy.
Figures


Similar articles
-
SUMOylation by SUMO2 is implicated in the degradation of misfolded ataxin-7 via RNF4 in SCA7 models.Dis Model Mech. 2019 Jan 11;12(1):dmm036145. doi: 10.1242/dmm.036145. Dis Model Mech. 2019. PMID: 30559154 Free PMC article.
-
SUMO-1 interacts with mutant ataxin-1 and colocalizes to its aggregates in Purkinje cells of SCA1 transgenic mice.Arch Ital Biol. 2010 Dec;148(4):351-63. doi: 10.4449/aib.v148i4.1201. Arch Ital Biol. 2010. PMID: 21308649
-
Polyglutamine-expanded ataxin-7 decreases nuclear translocation of NF-kappaB p65 and impairs NF-kappaB activity by inhibiting proteasome activity of cerebellar neurons.Cell Signal. 2007 Mar;19(3):573-81. doi: 10.1016/j.cellsig.2006.08.006. Epub 2006 Aug 23. Cell Signal. 2007. PMID: 17005371
-
Toward therapeutic targets for SCA3: Insight into the role of Machado-Joseph disease protein ataxin-3 in misfolded proteins clearance.Prog Neurobiol. 2015 Sep;132:34-58. doi: 10.1016/j.pneurobio.2015.06.004. Epub 2015 Jun 27. Prog Neurobiol. 2015. PMID: 26123252 Review.
-
Progress in pathogenesis studies of spinocerebellar ataxia type 1.Philos Trans R Soc Lond B Biol Sci. 1999 Jun 29;354(1386):1079-81. doi: 10.1098/rstb.1999.0462. Philos Trans R Soc Lond B Biol Sci. 1999. PMID: 10434309 Free PMC article. Review.
Cited by
-
Selective Autophagy by Close Encounters of the Ubiquitin Kind.Cells. 2020 Oct 24;9(11):2349. doi: 10.3390/cells9112349. Cells. 2020. PMID: 33114389 Free PMC article. Review.
-
Identifying Therapeutic Targets for Spinocerebellar Ataxia Type 3/Machado-Joseph Disease through Integration of Pathological Biomarkers and Therapeutic Strategies.Int J Mol Sci. 2020 Apr 26;21(9):3063. doi: 10.3390/ijms21093063. Int J Mol Sci. 2020. PMID: 32357546 Free PMC article. Review.
-
Crosstalk Between Mammalian Autophagy and the Ubiquitin-Proteasome System.Front Cell Dev Biol. 2018 Oct 2;6:128. doi: 10.3389/fcell.2018.00128. eCollection 2018. Front Cell Dev Biol. 2018. PMID: 30333975 Free PMC article. Review.
-
Mycobacteria modulate SUMOylation to suppresses protective responses in dendritic cells.PLoS One. 2023 Sep 29;18(9):e0283448. doi: 10.1371/journal.pone.0283448. eCollection 2023. PLoS One. 2023. PMID: 37773921 Free PMC article.
References
LinkOut - more resources
Full Text Sources
