Discovering RNA-Based Regulatory Systems for Yersinia Virulence
- PMID: 30460205
- PMCID: PMC6232918
- DOI: 10.3389/fcimb.2018.00378
Discovering RNA-Based Regulatory Systems for Yersinia Virulence
Abstract
The genus Yersinia includes three human pathogenic species, Yersinia pestis, the causative agent of the bubonic and pneumonic plague, and enteric pathogens Y. enterocolitica and Y. pseudotuberculosis that cause a number of gut-associated diseases. Over the past years a large repertoire of RNA-based regulatory systems has been discovered in these pathogens using different RNA-seq based approaches. Among them are several conserved or species-specific RNA-binding proteins, regulatory and sensory RNAs as well as various RNA-degrading enzymes. Many of them were shown to control the expression of important virulence-relevant factors and have a very strong impact on Yersinia virulence. The precise targets, the molecular mechanism and their role for Yersinia pathogenicity is only known for a small subset of identified genus- or species-specific RNA-based control elements. However, the ongoing development of new RNA-seq based methods and data analysis methods to investigate the synthesis, composition, translation, decay, and modification of RNAs in the bacterial cell will help us to generate a more comprehensive view of Yersinia RNA biology in the near future.
Keywords: Csr/Rsm system; RNA processing; RNA stability; RNA thermometer; gene regulation; small regulatory RNAs; virulence.
Figures
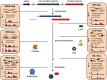
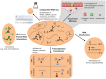
References
-
- Baba K., Takeda N., Tanaka M. (1991). Cases of Yersinia pseudotuberculosis infection having diagnostic criteria of Kawasaki disease. Contrib. Microbiol. Immunol. 12, 292–296. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

