BulkVis: a graphical viewer for Oxford nanopore bulk FAST5 files
- PMID: 30462145
- PMCID: PMC6596899
- DOI: 10.1093/bioinformatics/bty841
BulkVis: a graphical viewer for Oxford nanopore bulk FAST5 files
Abstract
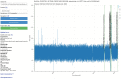
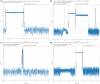
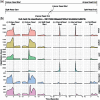
Motivation: The Oxford Nanopore Technologies (ONT) MinION is used for sequencing a wide variety of sample types with diverse methods of sample extraction. Nanopore sequencers output FAST5 files containing signal data subsequently base called to FASTQ format. Optionally, ONT devices can collect data from all sequencing channels simultaneously in a bulk FAST5 file enabling inspection of signal in any channel at any point. We sought to visualize this signal to inspect challenging or difficult to sequence samples.
Results: The BulkVis tool can load a bulk FAST5 file and overlays MinKNOW (the software that controls ONT sequencers) classifications on the signal trace and can show mappings to a reference. Users can navigate to a channel and time or, given a FASTQ header from a read, jump to its specific position. BulkVis can export regions as Nanopore base caller compatible reads. Using BulkVis, we find long reads can be incorrectly divided by MinKNOW resulting in single DNA molecules being split into two or more reads. The longest seen to date is 2 272 580 bases in length and reported in eleven consecutive reads. We provide helper scripts that identify and reconstruct split reads given a sequencing summary file and alignment to a reference. We note that incorrect read splitting appears to vary according to input sample type and is more common in 'ultra-long' read preparations.
Availability and implementation: The software is available freely under an MIT license at https://github.com/LooseLab/bulkvis.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2018. Published by Oxford University Press.
Figures

References
-
- Collette A. (2013) Python and HDF5. O’Reilly Media, Incorporated.
Publication types
MeSH terms
Substances
Grants and funding
- BB/M020061/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 204843/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- BB/N017099/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 1949454/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M008770/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

