Microglial phenotypes in the human epileptic temporal lobe
- PMID: 30462183
- PMCID: PMC6277010
- DOI: 10.1093/brain/awy276
Microglial phenotypes in the human epileptic temporal lobe
Erratum in
-
Corrigendum.Brain. 2020 Jan 1;143(1):e7. doi: 10.1093/brain/awz359. Brain. 2020. PMID: 31711112 No abstract available.
Abstract
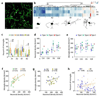
Microglia, the immune cells of the brain, are highly plastic and possess multiple functional phenotypes. Differences in phenotype in different regions and different states of epileptic human brain have been little studied. Here we use transcriptomics, anatomy, imaging of living cells and ELISA measurements of cytokine release to examine microglia from patients with temporal lobe epilepsies. Two distinct microglial phenotypes were explored. First we asked how microglial phenotype differs between regions of high and low neuronal loss in the same brain. Second, we asked how microglial phenotype is changed by a recent seizure. In sclerotic areas with few neurons, microglia have an amoeboid rather than ramified shape, express activation markers and respond faster to purinergic stimuli. The repairing interleukin, IL-10, regulates the basal phenotype of microglia in the CA1 and CA3 regions with neuronal loss and gliosis. To understand changes in phenotype induced by a seizure, we estimated the delay from the last seizure until tissue collection from changes in reads for immediate early gene transcripts. Pseudotime ordering of these data was validated by comparison with results from kainate-treated mice. It revealed a local and transient phenotype in which microglia secrete the human interleukin CXCL8, IL-1B and other cytokines. This secretory response is mediated in part via the NRLP3 inflammasome.
Conflict of interest statement
None of the authors have competing interests.
Figures






References
-
- Beach TG, Woodhurst WB, MacDonald DB, Jones MW. Reactive microglia in hippocampal sclerosis associated with human temporal lobe epilepsy. Neurosci Lett. 1995;191:27–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

