Dynamic transcriptomic analysis reveals suppression of PGC1α/ERRα drives perturbed myogenesis in facioscapulohumeral muscular dystrophy
- PMID: 30462217
- PMCID: PMC6452176
- DOI: 10.1093/hmg/ddy405
Dynamic transcriptomic analysis reveals suppression of PGC1α/ERRα drives perturbed myogenesis in facioscapulohumeral muscular dystrophy
Abstract
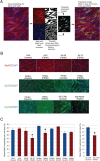
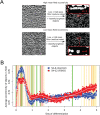
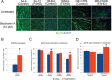
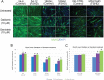
Facioscapulohumeral muscular dystrophy (FSHD) is a prevalent, incurable myopathy, linked to epigenetic derepression of D4Z4 repeats on chromosome 4q, leading to ectopic DUX4 expression. FSHD patient myoblasts have defective myogenic differentiation, forming smaller myotubes with reduced myosin content. However, molecular mechanisms driving such disrupted myogenesis in FSHD are poorly understood. We performed high-throughput morphological analysis describing FSHD and control myogenesis, revealing altered myogenic differentiation results in hypotrophic myotubes. Employing polynomial models and an empirical Bayes approach, we established eight critical time points during which human healthy and FSHD myogenesis differ. RNA-sequencing at these eight nodal time points in triplicate, provided temporal depth for a multivariate regression analysis, allowing assessment of interaction between progression of differentiation and FSHD disease status. Importantly, the unique size and structure of our data permitted identification of many novel FSHD pathomechanisms undetectable by previous approaches. For further analysis here, we selected pathways that control mitochondria: of interest considering known alterations in mitochondrial structure and function in FSHD muscle, and sensitivity of FSHD cells to oxidative stress. Notably, we identified suppression of mitochondrial biogenesis, in particular via peroxisome proliferator-activated receptor gamma coactivator 1-α (PGC1α), the cofactor and activator of oestrogen-related receptor α (ERRα). PGC1α knock-down caused hypotrophic myotubes to form from control myoblasts. Known ERRα agonists and safe food supplements biochanin A, daidzein or genistein, each rescued the hypotrophic FSHD myotube phenotype. Together our work describes transcriptomic changes in high resolution that occur during myogenesis in FSHD ex vivo, identifying suppression of the PGC1α-ERRα axis leading to perturbed myogenic differentiation, which can effectively be rescued by readily available food supplements.
© The Author(s) 2018. Published by Oxford University Press.
Figures



References
-
- Orrell R.W. (2011) Facioscapulohumeral dystrophy and scapuloperoneal syndromes. Handb Clin Neurol., 101, 167–180. - PubMed
-
- Osborne R.J., Welle S., Venance S.L., Thornton C.A. and Tawil R. (2007) Expression profile of FSHD supports a link between retinal vasculopathy and muscular dystrophy. Neurology, 68, 569–577. - PubMed
-
- Fitzsimons R.B. (2011) Retinal vascular disease and the pathogenesis of facioscapulohumeral muscular dystrophy. A signalling message from Wnt? Neuromuscul. Disord., 21, 263–271. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

