Inosine induces context-dependent recoding and translational stalling
- PMID: 30462291
- PMCID: PMC6326813
- DOI: 10.1093/nar/gky1163
Inosine induces context-dependent recoding and translational stalling
Abstract
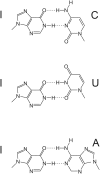
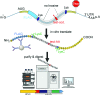
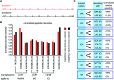
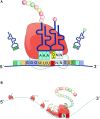
RNA modifications are present in all classes of RNAs. They control the fate of mRNAs by affecting their processing, translation, or stability. Inosine is a particularly widespread modification in metazoan mRNA arising from deamination of adenosine catalyzed by the RNA-targeting adenosine deaminases ADAR1 or ADAR2. Inosine is commonly thought to be interpreted as guanosine by cellular machines and during translation. Here, we systematically test ribosomal decoding using mass spectrometry. We show that while inosine is primarily interpreted as guanosine it can also be decoded as adenosine, and rarely even as uracil. Decoding of inosine as adenosine and uracil is context-dependent. In addition, mass spectrometry analysis indicates that inosine causes ribosome stalling especially when multiple inosines are present in the codon. Indeed, ribosome profiling data from human tissues confirm inosine-dependent ribosome stalling in vivo. To our knowledge this is the first study where decoding of inosine is tested in a comprehensive and unbiased way. Thus, our study shows novel, unanticipated functions for inosines in mRNAs, further expanding coding potential and affecting translational efficiency.
Figures



References
-
- Huang Y.S., Lu W.H.. Decoding hidden messages in neurons: insights from epitranscriptome-controlled and specialized ribosome-controlled translation. Curr. Opin. Neurobiol. 2018; 48:64–70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

