MBGD update 2018: microbial genome database based on hierarchical orthology relations covering closely related and distantly related comparisons
- PMID: 30462302
- PMCID: PMC6324027
- DOI: 10.1093/nar/gky1054
MBGD update 2018: microbial genome database based on hierarchical orthology relations covering closely related and distantly related comparisons
Abstract
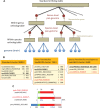
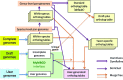
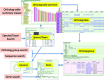
The Microbial Genome Database for Comparative Analysis (MBGD) is a database for comparative genomics based on comprehensive orthology analysis of bacteria, archaea and unicellular eukaryotes. MBGD now contains 6318 genomes. To utilize the database for both closely related and distantly related genomes, MBGD previously provided two types of ortholog tables: the standard ortholog table containing one representative genome from each genus covering the entire taxonomic range and the taxon specific ortholog tables for each taxon. However, this approach has a drawback in that the standard ortholog table contains only genes that are conserved in the representative genomes. To address this problem, we developed a stepwise procedure to construct ortholog tables hierarchically in a bottom-up manner. By using this approach, the new standard ortholog table now covers the entire gene repertoire stored in MBGD. In addition, we have enhanced several functionalities, including rapid and flexible keyword searching, profile-based sequence searching for orthology assignment to a user query sequence, and displaying a phylogenetic tree of each taxon based on the concatenated core gene sequences. For integrative database searching, the core data in MBGD are represented in Resource Description Framework (RDF) and a SPARQL interface is provided to search them. MBGD is available at http://mbgd.genome.ad.jp/.
Figures

References
-
- Tettelin H., Masignani V., Cieslewicz M.J., Donati C., Medini D., Ward N.L., Angiuoli S.V., Crabtree J., Jones A.L., Durkin A.S. et al. . Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:13950–13955. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

