Design of highly active double-pseudoknotted ribozymes: a combined computational and experimental study
- PMID: 30462314
- PMCID: PMC6326823
- DOI: 10.1093/nar/gky1118
Design of highly active double-pseudoknotted ribozymes: a combined computational and experimental study
Abstract
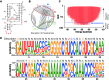
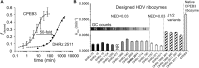
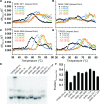
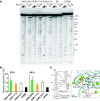
Design of RNA sequences that adopt functional folds establishes principles of RNA folding and applications in biotechnology. Inverse folding for RNAs, which allows computational design of sequences that adopt specific structures, can be utilized for unveiling RNA functions and developing genetic tools in synthetic biology. Although many algorithms for inverse RNA folding have been developed, the pseudoknot, which plays a key role in folding of ribozymes and riboswitches, is not addressed in most algorithms. For the few algorithms that attempt to predict pseudoknot-containing ribozymes, self-cleavage activity has not been tested. Herein, we design double-pseudoknot HDV ribozymes using an inverse RNA folding algorithm and test their kinetic mechanisms experimentally. More than 90% of the positively designed ribozymes possess self-cleaving activity, whereas more than 70% of negative control ribozymes, which are predicted to fold to the necessary structure but with low fidelity, do not possess it. Kinetic and mutation analyses reveal that these RNAs cleave site-specifically and with the same mechanism as the WT ribozyme. Most ribozymes react just 50- to 80-fold slower than the WT ribozyme, and this rate can be improved to near WT by modification of a junction. Thus, fast-cleaving functional ribozymes with multiple pseudoknots can be designed computationally.
Figures



Similar articles
-
Computer-Aided Design of Active Pseudoknotted Hammerhead Ribozymes.Methods Mol Biol. 2021;2167:91-111. doi: 10.1007/978-1-0716-0716-9_7. Methods Mol Biol. 2021. PMID: 32712917
-
Inverse RNA Folding Workflow to Design and Test Ribozymes that Include Pseudoknots.Methods Mol Biol. 2021;2167:113-143. doi: 10.1007/978-1-0716-0716-9_8. Methods Mol Biol. 2021. PMID: 32712918
-
Computational design and experimental verification of pseudoknotted ribozymes.RNA. 2023 Jun;29(6):764-776. doi: 10.1261/rna.079148.122. Epub 2023 Mar 3. RNA. 2023. PMID: 36868786 Free PMC article.
-
Catalytic RNA, ribozyme, and its applications in synthetic biology.Biotechnol Adv. 2019 Dec;37(8):107452. doi: 10.1016/j.biotechadv.2019.107452. Epub 2019 Oct 24. Biotechnol Adv. 2019. PMID: 31669138 Review.
-
HDV ribozymes.Curr Top Microbiol Immunol. 2006;307:47-65. doi: 10.1007/3-540-29802-9_3. Curr Top Microbiol Immunol. 2006. PMID: 16903220 Review.
Cited by
-
Cellular Concentrations of Nucleotide Diphosphate-Chelated Magnesium Ions Accelerate Catalysis by RNA and DNA Enzymes.Biochemistry. 2019 Sep 24;58(38):3971-3979. doi: 10.1021/acs.biochem.9b00578. Epub 2019 Sep 12. Biochemistry. 2019. PMID: 31512860 Free PMC article.
-
Rational design of oligonucleotides for enhanced in vitro transcription of small RNA.RNA. 2024 May 16;30(6):710-727. doi: 10.1261/rna.079923.123. RNA. 2024. PMID: 38423625 Free PMC article.
-
Predicting higher-order mutational effects in an RNA enzyme by machine learning of high-throughput experimental data.Front Mol Biosci. 2022 Aug 15;9:893864. doi: 10.3389/fmolb.2022.893864. eCollection 2022. Front Mol Biosci. 2022. PMID: 36046603 Free PMC article.
-
Strategies for improving the genome-editing efficiency of class 2 CRISPR/Cas system.Heliyon. 2024 Sep 27;10(19):e38588. doi: 10.1016/j.heliyon.2024.e38588. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39397905 Free PMC article. Review.
-
Sampling-based Continuous Optimization with Coupled Variables for RNA Design.ArXiv [Preprint]. 2024 Dec 11:arXiv:2412.08751v1. ArXiv. 2024. PMID: 39713799 Free PMC article. Preprint.
References
-
- Isaacs F.J., Dwyer D.J., Collins J.J.. RNA synthetic biology. Nat. Biotechnol. 2006; 24:545–554. - PubMed
-
- Chappell J., Watters K.E., Takahashi M.K., Lucks J.B.. A renaissance in RNA synthetic biology: new mechanisms, applications and tools for the future. Curr. Opin. Chem. Biol. 2015; 28:47–56. - PubMed
-
- Bartel D.P., Szostak J.W.. Isolation of new ribozymes from a large pool of random sequences. Science. 1993; 261:1411–1418. - PubMed
-
- Wilson D.S., Szostak J.W.. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999; 68:611–647. - PubMed
-
- Hofacker I.L., Fontana W., Stadler P.F., Bonhoeffer L.S., Tacker M., Schuster P.. Fast folding and comparison of RNA secondary structures. Monatshefte für Chem. 1994; 125:1434–4475.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

