The Molecular Signature of Megakaryocyte-Erythroid Progenitors Reveals a Role for the Cell Cycle in Fate Specification
- PMID: 30463007
- PMCID: PMC6336197
- DOI: 10.1016/j.celrep.2018.10.084
The Molecular Signature of Megakaryocyte-Erythroid Progenitors Reveals a Role for the Cell Cycle in Fate Specification
Erratum in
-
The Molecular Signature of Megakaryocyte-Erythroid Progenitors Reveals a Role for the Cell Cycle in Fate Specification.Cell Rep. 2018 Dec 11;25(11):3229. doi: 10.1016/j.celrep.2018.11.075. Cell Rep. 2018. PMID: 30540953 Free PMC article. No abstract available.
Abstract
Megakaryocytic-erythroid progenitors (MEPs) give rise to the cells that produce red blood cells and platelets. Although the mechanisms underlying megakaryocytic (MK) and erythroid (E) maturation have been described, those controlling their specification from MEPs are unknown. Single-cell RNA sequencing of primary human MEPs, common myeloid progenitors (CMPs), megakaryocyte progenitors, and E progenitors revealed a distinct transitional MEP signature. Inferred regulatory transcription factors (TFs) were associated with differential expression of cell cycle regulators. Genetic manipulation of selected TFs validated their role in lineage specification and demonstrated coincident modulation of the cell cycle. Genetic and pharmacologic modulation demonstrated that cell cycle activation is sufficient to promote E versus MK specification. These findings, obtained from healthy human cells, lay a foundation to study the mechanisms underlying benign and malignant disease states of the megakaryocytic and E lineages.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures

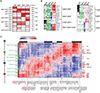


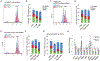

Comment in
-
Stepping Up Human Beige Fat Cell Production.Cell Rep. 2018 Dec 11;25(11):2935-2936. doi: 10.1016/j.celrep.2018.11.091. Cell Rep. 2018. PMID: 30540928
References
-
- Akashi K, Traver D, Miyamoto T, and Weissman IL (2000). A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404, 193–197. - PubMed
-
- Blackwood EM, and Eisenman RN (1991). Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science 251, 1211–1217. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

