Variant information systems for precision oncology
- PMID: 30463544
- PMCID: PMC6249891
- DOI: 10.1186/s12911-018-0665-z
Variant information systems for precision oncology
Abstract
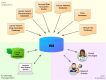
Background: The decreasing cost of obtaining high-quality calls of genomic variants and the increasing availability of clinically relevant data on such variants are important drivers for personalized oncology. To allow rational genome-based decisions in diagnosis and treatment, clinicians need intuitive access to up-to-date and comprehensive variant information, encompassing, for instance, prevalence in populations and diseases, functional impact at the molecular level, associations to druggable targets, or results from clinical trials. In practice, collecting such comprehensive information on genomic variants is difficult since the underlying data is dispersed over a multitude of distributed, heterogeneous, sometimes conflicting, and quickly evolving data sources. To work efficiently, clinicians require powerful Variant Information Systems (VIS) which automatically collect and aggregate available evidences from such data sources without suppressing existing uncertainty.
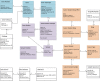
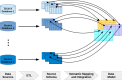
Methods: We address the most important cornerstones of modeling a VIS: We take from emerging community standards regarding the necessary breadth of variant information and procedures for their clinical assessment, long standing experience in implementing biomedical databases and information systems, our own clinical record of diagnosis and treatment of cancer patients based on molecular profiles, and extensive literature review to derive a set of design principles along which we develop a relational data model for variant level data. In addition, we characterize a number of public variant data sources, and describe a data integration pipeline to integrate their data into a VIS.
Results: We provide a number of contributions that are fundamental to the design and implementation of a comprehensive, operational VIS. In particular, we (a) present a relational data model to accurately reflect data extracted from public databases relevant for clinical variant interpretation, (b) introduce a fault tolerant and performant integration pipeline for public variant data sources, and (c) offer recommendations regarding a number of intricate challenges encountered when integrating variant data for clincal interpretation.
Conclusion: The analysis of requirements for representation of variant level data in an operational data model, together with the implementation-ready relational data model presented here, and the instructional description of methods to acquire comprehensive information to fill it, are an important step towards variant information systems for genomic medicine.
Keywords: Data model; Genomic variant data integration; Molecular cancer therapy; Variant information system.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Griffith M, Spies NC, Krysiak K, McMichael JF, Coffman AC, Danos AM, Ainscough BJ, Ramirez CA, Rieke DT, Kujan L, et al. Civic is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat Genet. 2017;49(2):170–4. doi: 10.1038/ng.3774. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

