Large-scale prediction of protein ubiquitination sites using a multimodal deep architecture
- PMID: 30463553
- PMCID: PMC6249717
- DOI: 10.1186/s12918-018-0628-0
Large-scale prediction of protein ubiquitination sites using a multimodal deep architecture
Abstract
Background: Ubiquitination, which is also called "lysine ubiquitination", occurs when an ubiquitin is attached to lysine (K) residues in targeting proteins. As one of the most important post translational modifications (PTMs), it plays the significant role not only in protein degradation, but also in other cellular functions. Thus, systematic anatomy of the ubiquitination proteome is an appealing and challenging research topic. The existing methods for identifying protein ubiquitination sites can be divided into two kinds: mass spectrometry and computational methods. Mass spectrometry-based experimental methods can discover ubiquitination sites from eukaryotes, but are time-consuming and expensive. Therefore, it is priority to develop computational approaches that can effectively and accurately identify protein ubiquitination sites.
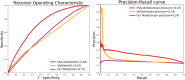
Results: The existing computational methods usually require feature engineering, which may lead to redundancy and biased representations. While deep learning is able to excavate underlying characteristics from large-scale training data via multiple-layer networks and non-linear mapping operations. In this paper, we proposed a deep architecture within multiple modalities to identify the ubiquitination sites. First, according to prior knowledge and biological knowledge, we encoded protein sequence fragments around candidate ubiquitination sites into three modalities, namely raw protein sequence fragments, physico-chemical properties and sequence profiles, and designed different deep network layers to extract the hidden representations from them. Then, the generative deep representations corresponding to three modalities were merged to build the final model. We performed our algorithm on the available largest scale protein ubiquitination sites database PLMD, and achieved 66.4% specificity, 66.7% sensitivity, 66.43% accuracy, and 0.221 MCC value. A number of comparative experiments also indicated that our multimodal deep architecture outperformed several popular protein ubiquitination site prediction tools.
Conclusion: The results of comparative experiments validated the effectiveness of our deep network and also displayed that our method outperformed several popular protein ubiquitination site prediction tools. The source codes of our proposed method are available at https://github.com/jiagenlee/deepUbiquitylation .
Keywords: Convolution neural network; Deep learning; Deep neural network; Multiple modalities; Protein ubiquitination site.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

