Genetic Analysis of NDT80 Family Transcription Factors in Candida albicans Using New CRISPR-Cas9 Approaches
- PMID: 30463924
- PMCID: PMC6249646
- DOI: 10.1128/mSphere.00545-18
Genetic Analysis of NDT80 Family Transcription Factors in Candida albicans Using New CRISPR-Cas9 Approaches
Abstract
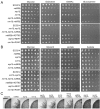
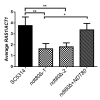
Ndt80 family transcription factors are highly conserved in fungi, where they regulate diverse processes. The human fungal pathogen Candida albicans contains three genes (NDT80, REP1, and RON1) that encode proteins with similarity to Saccharomyces cerevisiae Ndt80, although the homology is restricted to the DNA binding domain. To better understand their role in virulence functions, we used clustered regularly interspaced short palindromic repeat/CRISPR-associated gene 9 (CRISPR/Cas9) to delete the three NDT80-family genes. An ndt80Δ mutant showed strong defects in forming hyphae in response to serum or N-acetylglucosamine (GlcNAc), which was linked to the ability of Ndt80 to regulate the expression of RAS1, an upstream regulator of hyphal signaling. Conversely, the ndt80Δ mutant formed hyphal cells on glycerol medium, indicating that Ndt80 is not required for hyphal growth under all conditions. In contrast to our previously published data, a ron1Δ single mutant could grow and form hyphae in response to GlcNAc. However, deleting RON1 partially restored the ability of an ndt80Δ mutant to form hyphae in response to GlcNAc, indicating a link to GlcNAc signaling. REP1 was required for growth on GlcNAc, as expected, but not for GlcNAc or serum to induce hyphae. The ndt80Δ mutant was defective in growing under stressful conditions, such as elevated temperature, but not the ron1Δ mutant or rep1Δ mutant. Quantitative assays did not reveal any significant differences in the fluconazole susceptibility of the NDT80-family mutants. Interestingly, double and triple mutant analysis did not identify significant genetic interactions for these NDT80 family genes, indicating that they mainly function independently, in spite of their conserved DNA binding domain.IMPORTANCE Transcription factors play key roles in regulating virulence of the human fungal pathogen C. albicans In addition to regulating the expression of virulence factors, they also control the ability of C. albicans to switch to filamentous hyphal growth, which facilitates biofilm formation on medical devices and invasion into tissues. We therefore used new CRISPR/Cas9 methods to examine the effects of deleting three C. albicans genes (NDT80, REP1, and RON1) that encode transcription factors with similar DNA binding domains. Interestingly, double and triple mutant strains mostly showed the combined properties of the single mutants; there was only very limited evidence of synergistic interactions in regulating morphogenesis, stress resistance, and ability to metabolize different sugars. These results demonstrate that NDT80, REP1, and RON1 have distinct functions in regulating C. albicans virulence functions.
Keywords: Candida albicans; NDT80; REP1; RON1; hyphae; morphogenesis.
Copyright © 2018 Min et al.
Figures





Similar articles
-
N-acetylglucosamine Signaling: Transcriptional Dynamics of a Novel Sugar Sensing Cascade in a Model Pathogenic Yeast, Candida albicans.J Fungi (Basel). 2021 Jan 19;7(1):65. doi: 10.3390/jof7010065. J Fungi (Basel). 2021. PMID: 33477740 Free PMC article. Review.
-
Candida tropicalis RON1 is required for hyphal formation, biofilm development, and virulence but is dispensable for N-acetylglucosamine catabolism.Med Mycol. 2021 Apr 6;59(4):379-391. doi: 10.1093/mmy/myaa063. Med Mycol. 2021. PMID: 32712662
-
Regulation of Hyphal Growth and N-Acetylglucosamine Catabolism by Two Transcription Factors in Candida albicans.Genetics. 2017 May;206(1):299-314. doi: 10.1534/genetics.117.201491. Epub 2017 Mar 27. Genetics. 2017. PMID: 28348062 Free PMC article.
-
N-acetylglucosamine-mediated morphological transition in Candida albicans and Candida tropicalis.Curr Genet. 2021 Apr;67(2):249-254. doi: 10.1007/s00294-020-01138-z. Epub 2021 Jan 2. Curr Genet. 2021. PMID: 33388851 Review.
-
The N-acetylglucosamine catabolic gene cluster in Trichoderma reesei is controlled by the Ndt80-like transcription factor RON1.Mol Microbiol. 2016 Feb;99(4):640-57. doi: 10.1111/mmi.13256. Epub 2015 Nov 19. Mol Microbiol. 2016. PMID: 26481444 Free PMC article.
Cited by
-
N-acetylglucosamine Signaling: Transcriptional Dynamics of a Novel Sugar Sensing Cascade in a Model Pathogenic Yeast, Candida albicans.J Fungi (Basel). 2021 Jan 19;7(1):65. doi: 10.3390/jof7010065. J Fungi (Basel). 2021. PMID: 33477740 Free PMC article. Review.
-
Targeted Genetic Changes in Candida albicans Using Transient CRISPR-Cas9 Expression.Curr Protoc. 2021 Jan;1(1):e19. doi: 10.1002/cpz1.19. Curr Protoc. 2021. PMID: 33491919 Free PMC article.
-
Ten decadal advances in fungal biology leading towards human well-being.Fungal Divers. 2022;116(1):547-614. doi: 10.1007/s13225-022-00510-3. Epub 2022 Sep 15. Fungal Divers. 2022. PMID: 36123995 Free PMC article. Review.
-
Linking Cellular Morphogenesis with Antifungal Treatment and Susceptibility in Candida Pathogens.J Fungi (Basel). 2019 Feb 21;5(1):17. doi: 10.3390/jof5010017. J Fungi (Basel). 2019. PMID: 30795580 Free PMC article. Review.
-
Superior Conjugative Plasmids Delivered by Bacteria to Diverse Fungi.Biodes Res. 2022 Aug 19;2022:9802168. doi: 10.34133/2022/9802168. eCollection 2022. Biodes Res. 2022. PMID: 37850145 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

