Impact of Amoxicillin-Clavulanate followed by Autologous Fecal Microbiota Transplantation on Fecal Microbiome Structure and Metabolic Potential
- PMID: 30463925
- PMCID: PMC6249645
- DOI: 10.1128/mSphereDirect.00588-18
Impact of Amoxicillin-Clavulanate followed by Autologous Fecal Microbiota Transplantation on Fecal Microbiome Structure and Metabolic Potential
Abstract
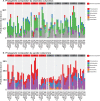
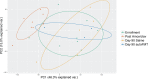
Strategies to prevent multidrug-resistant organism (MDRO) infections are scarce, but autologous fecal microbiota transplantation (autoFMT) may limit gastrointestinal MDRO expansion. AutoFMT involves banking one's feces during a healthy state for later use in restoring gut microbiota following perturbation. This pilot study evaluated the effect of autoFMT on gastrointestinal microbiome taxonomic composition, resistance gene content, and metabolic capacity after exposure to amoxicillin-clavulanic acid (Amox-Clav). Ten healthy participants were enrolled. All received 5 days of Amox-Clav. Half were randomized to autoFMT, derived from stool collected pre-antimicrobial exposure, by enema, and half to saline enema. Participants submitted stool samples pre- and post-Amox-Clav and enema and during a 90-day follow-up period. Shotgun metagenomic sequencing revealed taxonomic composition, resistance gene content, and metabolic capacity. Amox-Clav significantly altered gut taxonomic composition in all participants (n = 10, P < 0.01); however, only three participants exhibited major changes at the phylum level following exposure. In the cohort as a whole, beta-lactamase genes were enriched following Amox-Clav (P < 0.05), and predicted metabolic capacity was significantly altered (P < 0.01). Species composition, metabolic capacity, and beta-lactamase abundance returned to pre-antimicrobial exposure state 7 days after either autoFMT or saline enema (P > 0.05, compared to enrollment). Alterations to microbial metabolic capacity occurred following antimicrobial exposure even in participants without substantial taxonomic disruption, potentially creating open niches for pathogen colonization. Our findings suggest that metabolic potential is an important consideration for complete assessment of antimicrobial impact on the microbiome. AutoFMT was well tolerated and may have contributed to phylogenetic recovery. (This study has been registered at ClinicalTrials.gov under identifier NCT02046525.)IMPORTANCE The spread of multidrug resistance among pathogenic organisms threatens the efficacy of antimicrobial treatment options. The human gut serves as a reservoir for many drug-resistant organisms and their resistance genes, and perturbation of the gut microbiome by antimicrobial exposure can open metabolic niches to resistant pathogens. Once established in the gut, antimicrobial-resistant bacteria can persist even after antimicrobial exposure ceases. Strategies to prevent multidrug-resistant organism (MDRO) infections are scarce, but autologous fecal microbiota transplantation (autoFMT) may limit gastrointestinal MDRO expansion. AutoFMT involves banking one's feces during a healthy state for later use in restoring gut microbiota following perturbation. This pilot study evaluated the effect of amoxicillin-clavulanic acid (Amox-Clav) exposure and autoFMT on gastrointestinal microbiome taxonomic composition, resistance gene content, and metabolic capacity. Importantly, we found that metabolic capacity was perturbed even in cases where gross phylogeny remained unchanged and that autoFMT was safe and well tolerated.
Keywords: antimicrobial resistance; fecal microbiota transplantation; metagenomics; microbiome; multidrug resistance.
Copyright © 2018 Bulow et al.
Figures



Similar articles
-
Microbiota restoration reduces antibiotic-resistant bacteria gut colonization in patients with recurrent Clostridioides difficile infection from the open-label PUNCH CD study.Genome Med. 2021 Feb 16;13(1):28. doi: 10.1186/s13073-021-00843-9. Genome Med. 2021. PMID: 33593430 Free PMC article. Clinical Trial.
-
One dose ceftriaxone vs. ten days of amoxicillin/clavulanate therapy for acute otitis media: clinical efficacy and change in nasopharyngeal flora.Pediatr Infect Dis J. 1999 May;18(5):403-9. doi: 10.1097/00006454-199905000-00002. Pediatr Infect Dis J. 1999. PMID: 10353511 Clinical Trial.
-
Efficacy and safety of cefdinir in the treatment of maxillary sinusitis.Eur Arch Otorhinolaryngol. 2000;257(3):140-8. doi: 10.1007/s004050050211. Eur Arch Otorhinolaryngol. 2000. PMID: 10839487 Clinical Trial.
-
The role of the gut microbiome in colonization resistance and recurrent Clostridioides difficile infection.Therap Adv Gastroenterol. 2022 Nov 18;15:17562848221134396. doi: 10.1177/17562848221134396. eCollection 2022. Therap Adv Gastroenterol. 2022. PMID: 36425405 Free PMC article. Review.
-
Exploring the role of gut microbiota in antibiotic resistance and prevention.Ann Med. 2025 Dec;57(1):2478317. doi: 10.1080/07853890.2025.2478317. Epub 2025 Mar 17. Ann Med. 2025. PMID: 40096354 Free PMC article. Review.
Cited by
-
Autologous fecal microbiota transplantation for the treatment of inflammatory bowel disease.Transl Res. 2020 Dec;226:1-11. doi: 10.1016/j.trsl.2020.05.008. Epub 2020 Jun 22. Transl Res. 2020. PMID: 32585148 Free PMC article. Review.
-
Related Factor Analysis and Nursing Strategies of Diarrhea in Critically Ill Patients with Enteral Nutrition.Emerg Med Int. 2022 Sep 21;2022:8423048. doi: 10.1155/2022/8423048. eCollection 2022. Emerg Med Int. 2022. Retraction in: Emerg Med Int. 2023 Nov 29;2023:9803698. doi: 10.1155/2023/9803698. PMID: 36186529 Free PMC article. Retracted.
-
Fecal Microbial Transplantation and Its Potential Application in Cardiometabolic Syndrome.Front Immunol. 2019 Jun 14;10:1341. doi: 10.3389/fimmu.2019.01341. eCollection 2019. Front Immunol. 2019. PMID: 31258528 Free PMC article. Review.
-
Understanding the impact of antibiotic perturbation on the human microbiome.Genome Med. 2020 Sep 28;12(1):82. doi: 10.1186/s13073-020-00782-x. Genome Med. 2020. PMID: 32988391 Free PMC article. Review.
-
Machine learning analysis to identify the association between risk factors and onset of nosocomial diarrhea: a retrospective cohort study.PeerJ. 2019 Oct 30;7:e7969. doi: 10.7717/peerj.7969. eCollection 2019. PeerJ. 2019. PMID: 31687281 Free PMC article.
References
-
- World Health Organization. 2014. Antimicrobial resistance: global report on surveillance. World Health Organization, Geneva, Switzerland.
-
- Munoz-Price LS, De La Cuesta C, Adams S, Wyckoff M, Cleary T, McCurdy SP, Huband MD, Lemmon MM, Lescoe M, Dibhajj FB, Hayden MK, Lolans K, Quinn JP. 2010. Successful eradication of a monoclonal strain of Klebsiella pneumoniae during a K. pneumoniae carbapenemase-producing K. pneumoniae outbreak in a surgical intensive care unit in Miami, Florida. Infect Control Hosp Epidemiol 31:1074–1077. doi:10.1086/656243. - DOI - PubMed
-
- Bhalla A, Pultz NJ, Ray AJ, Hoyen CK, Eckstein EC, Donskey CJ. 2003. Antianaerobic antibiotic therapy promotes overgrowth of antibiotic-resistant, gram-negative bacilli and vancomycin-resistant enterococci in the stool of colonized patients. Infect Control Hosp Epidemiol 24:644–649. doi:10.1086/502267. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

