The calculation of transcript flux ratios reveals single regulatory mechanisms capable of activation and repression
- PMID: 30463953
- PMCID: PMC6294943
- DOI: 10.1073/pnas.1809454115
The calculation of transcript flux ratios reveals single regulatory mechanisms capable of activation and repression
Abstract
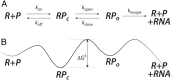
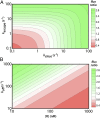
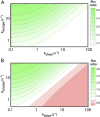
The regulation of transcription allows cells to adjust the rate of RNA polymerases (RNAPs) initiated in a promoter-specific manner. Classically, transcription factors are directed to a subset of promoters via the recognition of DNA sequence motifs. However, a unique class of regulators is recruited directly through interactions with RNAP. Surprisingly, these factors may still possess promoter specificity, and it has been postulated that the same kinetic mechanism leads to different regulatory outcomes depending on a promoter's basal rate constants. However, mechanistic studies of regulation typically report factor activity in terms of changes in the thermodynamics or kinetics of individual steps or states while qualitatively linking these observations to measured changes in transcript production. Here, I present online calculators that allow for the direct testing of mechanistic hypotheses by calculating the steady-state transcript flux in the presence and absence of a factor as a function of initiation rate constants. By evaluating how the flux ratio of a single kinetic mechanism varies across promoter space, quantitative insights into the potential of a mechanism to generate promoter-specific regulatory outcomes are obtained. Using these calculations, I predict that the mycobacterial transcription factor CarD is capable of repression in addition to its known role as an activator of ribosomal genes. In addition, a modification of the mechanism of the stringent response factors DksA/guanosine 5'-diphosphate 3'-diphosphate (ppGpp) is proposed based on their ability to differentially regulate transcription across promoter space. Overall, I conclude that a multifaceted kinetic mechanism is a requirement for differential regulation by this class of factors.
Keywords: CarD; DksA; kinetics; regulation; transcription.
Conflict of interest statement
The author declares no conflict of interest.
Figures



Similar articles
-
CarD and RbpA modify the kinetics of initial transcription and slow promoter escape of the Mycobacterium tuberculosis RNA polymerase.Nucleic Acids Res. 2019 Jul 26;47(13):6685-6698. doi: 10.1093/nar/gkz449. Nucleic Acids Res. 2019. PMID: 31127308 Free PMC article.
-
Transcription regulation by CarD in mycobacteria is guided by basal promoter kinetics.J Biol Chem. 2023 Jun;299(6):104724. doi: 10.1016/j.jbc.2023.104724. Epub 2023 Apr 17. J Biol Chem. 2023. PMID: 37075846 Free PMC article.
-
CarD stabilizes mycobacterial open complexes via a two-tiered kinetic mechanism.Nucleic Acids Res. 2015 Mar 31;43(6):3272-85. doi: 10.1093/nar/gkv078. Epub 2015 Feb 19. Nucleic Acids Res. 2015. PMID: 25697505 Free PMC article.
-
Transcriptional Responses to ppGpp and DksA.Annu Rev Microbiol. 2018 Sep 8;72:163-184. doi: 10.1146/annurev-micro-090817-062444. Annu Rev Microbiol. 2018. PMID: 30200857 Free PMC article. Review.
-
Strength and regulation without transcription factors: lessons from bacterial rRNA promoters.Cold Spring Harb Symp Quant Biol. 1998;63:131-9. doi: 10.1101/sqb.1998.63.131. Cold Spring Harb Symp Quant Biol. 1998. PMID: 10384277 Review. No abstract available.
Cited by
-
High-throughput, fluorescent-aptamer-based measurements of steady-state transcription rates for the Mycobacterium tuberculosis RNA polymerase.Nucleic Acids Res. 2023 Oct 27;51(19):e99. doi: 10.1093/nar/gkad761. Nucleic Acids Res. 2023. PMID: 37739412 Free PMC article.
-
Regulation of steady state ribosomal transcription in Mycobacterium tuberculosis: Intersection of sigma subunits, superhelicity, and transcription factors.J Biol Chem. 2025 Jun 12;301(8):110369. doi: 10.1016/j.jbc.2025.110369. Online ahead of print. J Biol Chem. 2025. PMID: 40516872 Free PMC article.
-
Transcription factor interactions explain the context-dependent activity of CRX binding sites.PLoS Comput Biol. 2024 Jan 16;20(1):e1011802. doi: 10.1371/journal.pcbi.1011802. eCollection 2024 Jan. PLoS Comput Biol. 2024. PMID: 38227575 Free PMC article.
-
Diverse molecular mechanisms of transcription regulation by the bacterial alarmone ppGpp.Mol Microbiol. 2022 Feb;117(2):252-260. doi: 10.1111/mmi.14860. Epub 2021 Dec 25. Mol Microbiol. 2022. PMID: 34894005 Free PMC article. Review.
-
CarD and RbpA modify the kinetics of initial transcription and slow promoter escape of the Mycobacterium tuberculosis RNA polymerase.Nucleic Acids Res. 2019 Jul 26;47(13):6685-6698. doi: 10.1093/nar/gkz449. Nucleic Acids Res. 2019. PMID: 31127308 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

