Germline genetic polymorphisms influence tumor gene expression and immune cell infiltration
- PMID: 30463956
- PMCID: PMC6294879
- DOI: 10.1073/pnas.1804506115
Germline genetic polymorphisms influence tumor gene expression and immune cell infiltration
Abstract
Cancer immunotherapy has emerged as an effective therapy in a variety of cancers. However, a key challenge in the field is that only a subset of patients who receive immunotherapy exhibit durable response. It has been hypothesized that host genetics influences the inherent immune profiles of patients and may underlie their differential response to immunotherapy. Herein, we systematically determined the association of common germline genetic variants with gene expression and immune cell infiltration of the tumor. We identified 64,094 expression quantitative trait loci (eQTLs) that associated with 18,210 genes (eGenes) across 24 human cancers. Overall, eGenes were enriched for their being involved in immune processes, suggesting that expression of immune genes can be shaped by hereditary genetic variants. We identified the endoplasmic reticulum aminopeptidase 2 (ERAP2) gene as a pan-cancer type eGene whose expression levels stratified overall survival in a subset of patients with bladder cancer receiving anti-PD-L1 (atezolizumab) therapy. Finally, we identified 103 gene signature QTLs (gsQTLs) that were associated with predicted immune cell abundance within the tumor microenvironment. Our findings highlight the impact of germline SNPs on cancer-immune phenotypes and response to therapy; and these analyses provide a resource for integration of germline genetics as a component of personalized cancer immunotherapy.
Keywords: TCGA; cancer immunology; eGenes; eQTL.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
Conflict of interest statement: All authors are employees of Genentech, Inc.
Figures

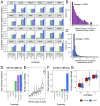
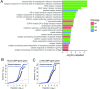
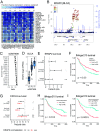

Comment in
-
Your genomic inheritance matters.Nat Rev Cancer. 2019 Feb;19(2):63. doi: 10.1038/s41568-018-0099-z. Nat Rev Cancer. 2019. PMID: 30578413 No abstract available.
Similar articles
-
Germline genetic contribution to the immune landscape of cancer.Immunity. 2021 Feb 9;54(2):367-386.e8. doi: 10.1016/j.immuni.2021.01.011. Immunity. 2021. PMID: 33567262 Free PMC article.
-
Association of Germline Variants in Natural Killer Cells With Tumor Immune Microenvironment Subtypes, Tumor-Infiltrating Lymphocytes, Immunotherapy Response, Clinical Outcomes, and Cancer Risk.JAMA Netw Open. 2019 Sep 4;2(9):e199292. doi: 10.1001/jamanetworkopen.2019.9292. JAMA Netw Open. 2019. PMID: 31483464 Free PMC article.
-
CancerImmunityQTL: a database to systematically evaluate the impact of genetic variants on immune infiltration in human cancer.Nucleic Acids Res. 2021 Jan 8;49(D1):D1065-D1073. doi: 10.1093/nar/gkaa805. Nucleic Acids Res. 2021. PMID: 33010176 Free PMC article.
-
Emerging role of immunotherapy in urothelial carcinoma-Immunobiology/biomarkers.Urol Oncol. 2016 Dec;34(12):556-565. doi: 10.1016/j.urolonc.2016.10.006. Epub 2016 Nov 9. Urol Oncol. 2016. PMID: 27836246 Free PMC article. Review.
-
Classifying Cancers Based on T-cell Infiltration and PD-L1.Cancer Res. 2015 Jun 1;75(11):2139-45. doi: 10.1158/0008-5472.CAN-15-0255. Cancer Res. 2015. PMID: 25977340 Free PMC article. Review.
Cited by
-
Common allotypes of ER aminopeptidase 1 have substrate-dependent and highly variable enzymatic properties.J Biol Chem. 2021 Jan-Jun;296:100443. doi: 10.1016/j.jbc.2021.100443. Epub 2021 Feb 20. J Biol Chem. 2021. PMID: 33617882 Free PMC article.
-
Antigen presentation in cancer: insights into tumour immunogenicity and immune evasion.Nat Rev Cancer. 2021 May;21(5):298-312. doi: 10.1038/s41568-021-00339-z. Epub 2021 Mar 9. Nat Rev Cancer. 2021. PMID: 33750922 Review.
-
A Comparative Review of Pregnancy and Cancer and Their Association with Endoplasmic Reticulum Aminopeptidase 1 and 2.Int J Mol Sci. 2023 Feb 9;24(4):3454. doi: 10.3390/ijms24043454. Int J Mol Sci. 2023. PMID: 36834865 Free PMC article. Review.
-
Germline polygenic risk scores are associated with immune gene expression signature and immune cell infiltration in breast cancer.Am J Hum Genet. 2024 Oct 3;111(10):2150-2163. doi: 10.1016/j.ajhg.2024.08.009. Epub 2024 Sep 12. Am J Hum Genet. 2024. PMID: 39270649 Free PMC article.
-
Does the gut microbiome environment influence response to systemic breast cancer treatment?Explor Target Antitumor Ther. 2021;2(4):374-384. doi: 10.37349/etat.2021.00051. Epub 2021 Aug 30. Explor Target Antitumor Ther. 2021. PMID: 36046753 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

