Structural snapshots of OxyR reveal the peroxidatic mechanism of H2O2 sensing
- PMID: 30463959
- PMCID: PMC6294878
- DOI: 10.1073/pnas.1807954115
Structural snapshots of OxyR reveal the peroxidatic mechanism of H2O2 sensing
Abstract
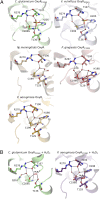
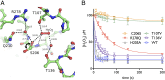
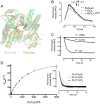
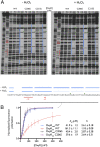
Hydrogen peroxide (H2O2) is a strong oxidant capable of oxidizing cysteinyl thiolates, yet only a few cysteine-containing proteins have exceptional reactivity toward H2O2 One such example is the prokaryotic transcription factor OxyR, which controls the antioxidant response in bacteria, and which specifically and rapidly reduces H2O2 In this study, we present crystallographic evidence for the H2O2-sensing mechanism and H2O2-dependent structural transition of Corynebacterium glutamicum OxyR by capturing the reduced and H2O2-bound structures of a serine mutant of the peroxidatic cysteine, and the full-length crystal structure of disulfide-bonded oxidized OxyR. In the H2O2-bound structure, we pinpoint the key residues for the peroxidatic reduction of H2O2, and relate this to mutational assays showing that the conserved active-site residues T107 and R278 are critical for effective H2O2 reduction. Furthermore, we propose an allosteric mode of structural change, whereby a localized conformational change arising from H2O2-induced intramolecular disulfide formation drives a structural shift at the dimerization interface of OxyR, leading to overall changes in quaternary structure and an altered DNA-binding topology and affinity at the catalase promoter region. This study provides molecular insights into the overall OxyR transcription mechanism regulated by H2O2.
Keywords: X-ray structure; hydrogen peroxide sensor; redox regulation; transcription factor.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
OxyR acts as a transcriptional repressor of hydrogen peroxide-inducible antioxidant genes in Corynebacterium glutamicum R.FEBS J. 2013 Jul;280(14):3298-312. doi: 10.1111/febs.12312. Epub 2013 Jun 3. FEBS J. 2013. PMID: 23621709
-
Structural details of the OxyR peroxide-sensing mechanism.Proc Natl Acad Sci U S A. 2015 May 19;112(20):6443-8. doi: 10.1073/pnas.1424495112. Epub 2015 Apr 30. Proc Natl Acad Sci U S A. 2015. PMID: 25931525 Free PMC article.
-
The hydrogen peroxide hypersensitivity of OxyR2 in Vibrio vulnificus depends on conformational constraints.J Biol Chem. 2017 Apr 28;292(17):7223-7232. doi: 10.1074/jbc.M116.743765. Epub 2017 Mar 6. J Biol Chem. 2017. PMID: 28264933 Free PMC article.
-
Redox-operated genetic switches: the SoxR and OxyR transcription factors.Trends Biotechnol. 2001 Mar;19(3):109-14. doi: 10.1016/s0167-7799(00)01542-0. Trends Biotechnol. 2001. PMID: 11179804 Review.
-
Hydrogen peroxide sensing, signaling and regulation of transcription factors.Redox Biol. 2014 Feb 23;2:535-62. doi: 10.1016/j.redox.2014.02.006. eCollection 2014. Redox Biol. 2014. PMID: 24634836 Free PMC article. Review.
Cited by
-
The reduced form of the antibody CH2 domain.Protein Sci. 2021 Sep;30(9):1895-1903. doi: 10.1002/pro.4142. Epub 2021 Jun 16. Protein Sci. 2021. PMID: 34107549 Free PMC article.
-
Selenium-Catalyzed Reduction of Hydroperoxides in Chemistry and Biology.Antioxidants (Basel). 2021 Sep 30;10(10):1560. doi: 10.3390/antiox10101560. Antioxidants (Basel). 2021. PMID: 34679695 Free PMC article. Review.
-
Prdx1 Interacts with ASK1 upon Exposure to H2O2 and Independently of a Scaffolding Protein.Antioxidants (Basel). 2021 Jun 30;10(7):1060. doi: 10.3390/antiox10071060. Antioxidants (Basel). 2021. PMID: 34209102 Free PMC article.
-
Bacterial Metallostasis: Metal Sensing, Metalloproteome Remodeling, and Metal Trafficking.Chem Rev. 2024 Dec 25;124(24):13574-13659. doi: 10.1021/acs.chemrev.4c00264. Epub 2024 Dec 10. Chem Rev. 2024. PMID: 39658019 Free PMC article. Review.
-
How Microbes Defend Themselves From Incoming Hydrogen Peroxide.Front Immunol. 2021 Apr 27;12:667343. doi: 10.3389/fimmu.2021.667343. eCollection 2021. Front Immunol. 2021. PMID: 33995399 Free PMC article. Review.
References
-
- Winterbourn CC. The biological chemistry of hydrogen peroxide. Methods Enzymol. 2013;528:3–25. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

