Existing Host Range Mutations Constrain Further Emergence of RNA Viruses
- PMID: 30463962
- PMCID: PMC6364021
- DOI: 10.1128/JVI.01385-18
Existing Host Range Mutations Constrain Further Emergence of RNA Viruses
Abstract
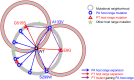
RNA viruses are capable of rapid host shifting, typically due to a point mutation that confers expanded host range. As additional point mutations are necessary for further expansions, epistasis among host range mutations can potentially affect the mutational neighborhood and frequency of niche expansion. We mapped the mutational neighborhood of host range expansion using three genotypes of the double-stranded RNA (dsRNA) bacteriophage φ6 (wild type and two isogenic host range mutants) on the novel host Pseudomonas syringae pv. atrofaciens. Both Sanger sequencing of 50 P. syringae pv. atrofaciens mutant clones for each genotype and population Illumina sequencing revealed the same high-frequency mutations allowing infection of P. syringae pv. atrofaciens. Wild-type φ6 had at least nine different ways of mutating to enter the novel host, eight of which are in p3 (host attachment protein gene), and 13/50 clones had unchanged p3 genes. However, the two isogenic mutants had dramatically restricted neighborhoods: only one or two mutations, all in p3. Deep sequencing revealed that wild-type clones without mutations in p3 likely had changes in p12 (morphogenic protein), a region that was not polymorphic for the two isogenic host range mutants. Sanger sequencing confirmed that 10/13 of the wild-type φ6 clones had nonsynonymous mutations in p12, and 2 others had point mutations in p9 and p5. None of these genes had previously been associated with host range expansion in φ6. We demonstrate, for the first time, epistatic constraint in an RNA virus due to host range mutations themselves, which has implications for models of serial host range expansion.IMPORTANCE RNA viruses mutate rapidly and frequently expand their host ranges to infect novel hosts, leading to serial host shifts. Using an RNA bacteriophage model system (Pseudomonas phage φ6), we studied the impact of preexisting host range mutations on another host range expansion. Results from both clonal Sanger and Illumina sequencing show that extant host range mutations dramatically narrow the neighborhood of potential host range mutations compared to that of wild-type φ6. This research suggests that serial host-shifting viruses may follow a small number of molecular paths to enter additional novel hosts. We also identified new genes involved in φ6 host range expansion, expanding our knowledge of this important model system in experimental evolution.
Keywords: RNA virus; entropy; epistasis; host range mutations.
Copyright © 2019 American Society for Microbiology.
Figures
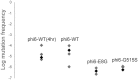
Similar articles
-
Experimental Evolution Studies in Φ6 Cystovirus.Viruses. 2024 Jun 18;16(6):977. doi: 10.3390/v16060977. Viruses. 2024. PMID: 38932268 Free PMC article. Review.
-
Pleiotropic costs of niche expansion in the RNA bacteriophage phi 6.Genetics. 2006 Feb;172(2):751-7. doi: 10.1534/genetics.105.051136. Epub 2005 Nov 19. Genetics. 2006. PMID: 16299384 Free PMC article.
-
High frequency of mutations that expand the host range of an RNA virus.Genetics. 2007 Jun;176(2):1013-22. doi: 10.1534/genetics.106.064634. Epub 2007 Apr 3. Genetics. 2007. PMID: 17409090 Free PMC article.
-
Frequency and fitness consequences of bacteriophage φ6 host range mutations.PLoS One. 2014 Nov 19;9(11):e113078. doi: 10.1371/journal.pone.0113078. eCollection 2014. PLoS One. 2014. PMID: 25409341 Free PMC article.
-
Structural Studies of Bacteriophage Φ6 and Its Transformations during Its Life Cycle.Viruses. 2023 Dec 11;15(12):2404. doi: 10.3390/v15122404. Viruses. 2023. PMID: 38140645 Free PMC article. Review.
Cited by
-
Viruses in astrobiology.Front Microbiol. 2022 Oct 26;13:1032918. doi: 10.3389/fmicb.2022.1032918. eCollection 2022. Front Microbiol. 2022. PMID: 36386652 Free PMC article. Review.
-
Competition-driven eco-evolutionary feedback reshapes bacteriophage lambda's fitness landscape and enables speciation.Nat Commun. 2024 Jan 29;15(1):863. doi: 10.1038/s41467-024-45008-5. Nat Commun. 2024. PMID: 38286804 Free PMC article.
-
Emerging roles of extracellular vesicles in mediating RNA virus infection.Fundam Res. 2021 Mar;1(2):179-185. doi: 10.1016/j.fmre.2021.02.005. Epub 2021 Feb 26. Fundam Res. 2021. PMID: 40477261 Free PMC article. Review.
-
Experimental Evolution Studies in Φ6 Cystovirus.Viruses. 2024 Jun 18;16(6):977. doi: 10.3390/v16060977. Viruses. 2024. PMID: 38932268 Free PMC article. Review.
-
Gauging genetic diversity of generalists: A test of genetic and ecological generalism with RNA virus experimental evolution.Virus Evol. 2019 Jun 29;5(1):vez019. doi: 10.1093/ve/vez019. eCollection 2019 Jan. Virus Evol. 2019. PMID: 31275611 Free PMC article.
References
-
- Marston HD, Folkers GK, Morens DM, Fauci AS. 2014. Emerging viral diseases: confronting threats with new technologies. Sci Transl Med 6:253ps210. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

