The Alphavirus E2 Membrane-Proximal Domain Impacts Capsid Interaction and Glycoprotein Lattice Formation
- PMID: 30463969
- PMCID: PMC6364017
- DOI: 10.1128/JVI.01881-18
The Alphavirus E2 Membrane-Proximal Domain Impacts Capsid Interaction and Glycoprotein Lattice Formation
Abstract
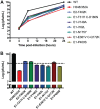
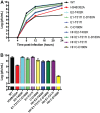
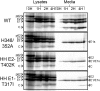
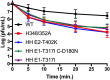
Alphaviruses are small enveloped RNA viruses that bud from the host cell plasma membrane. Alphavirus particles have a highly organized structure, with a nucleocapsid core containing the RNA genome surrounded by the capsid protein, and a viral envelope containing 80 spikes, each a trimer of heterodimers of the E1 and E2 glycoproteins. The capsid protein and envelope proteins are both arranged in organized lattices that are linked via the interaction of the E2 cytoplasmic tail/endodomain with the capsid protein. We previously characterized the role of two highly conserved histidine residues, H348 and H352, located in an external, juxtamembrane region of the E2 protein termed the D-loop. Alanine substitutions of H348 and H352 inhibit virus growth by impairing late steps in the assembly/budding of virus particles at the plasma membrane. To investigate this budding defect, we selected for revertants of the E2-H348/352A double mutant. We identified eleven second-site revertants with improved virus growth and mutations in the capsid, E2 and E1 proteins. Multiple isolates contained the mutation E2-T402K in the E2 endodomain or E1-T317I in the E1 ectodomain. Both of these mutations were shown to partially restore H348/352A growth and virus assembly/budding, while neither rescued the decreased thermostability of H348/352A. Within the alphavirus particle, these mutations are positioned to affect the E2-capsid interaction or the E1-mediated intertrimer interactions at the 5-fold axis of symmetry. Together, our results support a model in which the E2 D-loop promotes the formation of the glycoprotein lattice and its interactions with the internal capsid protein lattice.IMPORTANCE Alphaviruses include important human pathogens such as Chikungunya and the encephalitic alphaviruses. There are currently no licensed alphavirus vaccines or effective antiviral therapies, and more molecular information on virus particle structure and function is needed. Here, we highlight the important role of the E2 juxtamembrane D-loop in mediating virus budding and particle production. Our results demonstrated that this E2 region affects both the formation of the external glycoprotein lattice and its interactions with the internal capsid protein shell.
Keywords: alphavirus; virus assembly; virus budding.
Copyright © 2019 American Society for Microbiology.
Figures



Similar articles
-
An Alphavirus E2 Membrane-Proximal Domain Promotes Envelope Protein Lateral Interactions and Virus Budding.mBio. 2017 Nov 7;8(6):e01564-17. doi: 10.1128/mBio.01564-17. mBio. 2017. PMID: 29114027 Free PMC article.
-
Imaging the alphavirus exit pathway.J Virol. 2014 Jun;88(12):6922-33. doi: 10.1128/JVI.00592-14. Epub 2014 Apr 2. J Virol. 2014. PMID: 24696489 Free PMC article.
-
Mutations at the Alphavirus E1'-E2 Interdimer Interface Have Host-Specific Phenotypes.J Virol. 2022 Mar 9;96(5):e0214921. doi: 10.1128/jvi.02149-21. Epub 2022 Jan 12. J Virol. 2022. PMID: 35019719 Free PMC article.
-
Budding of alphaviruses.Virus Res. 2004 Dec;106(2):103-16. doi: 10.1016/j.virusres.2004.08.008. Virus Res. 2004. PMID: 15567491 Review.
-
The Alphavirus Exit Pathway: What We Know and What We Wish We Knew.Viruses. 2018 Feb 22;10(2):89. doi: 10.3390/v10020089. Viruses. 2018. PMID: 29470397 Free PMC article. Review.
Cited by
-
A Naturally Occurring Polymorphism in the Base of Sudan Virus Glycoprotein Decreases Glycoprotein Stability in a Species-Dependent Manner.J Virol. 2021 Aug 25;95(18):e0107321. doi: 10.1128/JVI.01073-21. Epub 2021 Aug 25. J Virol. 2021. PMID: 34232742 Free PMC article.
-
Structural constraints link differences in neutralization potency of human anti-Eastern equine encephalitis virus monoclonal antibodies.Proc Natl Acad Sci U S A. 2023 Mar 28;120(13):e2213690120. doi: 10.1073/pnas.2213690120. Epub 2023 Mar 24. Proc Natl Acad Sci U S A. 2023. PMID: 36961925 Free PMC article.
-
A molecular understanding of alphavirus entry.PLoS Pathog. 2020 Oct 22;16(10):e1008876. doi: 10.1371/journal.ppat.1008876. eCollection 2020 Oct. PLoS Pathog. 2020. PMID: 33091085 Free PMC article. Review.
-
Emerging Chikungunya Virus Variants at the E1-E1 Interglycoprotein Spike Interface Impact Virus Attachment and Inflammation.J Virol. 2022 Feb 23;96(4):e0158621. doi: 10.1128/JVI.01586-21. Epub 2021 Dec 22. J Virol. 2022. PMID: 34935436 Free PMC article.
-
Therapeutic alphavirus cross-reactive E1 human antibodies inhibit viral egress.Cell. 2021 Aug 19;184(17):4430-4446.e22. doi: 10.1016/j.cell.2021.07.033. Cell. 2021. PMID: 34416147 Free PMC article.
References
-
- Kuhn RJ. 2013. Togaviridae, p 629–650. In Knipe DM, Howley PM (ed), Fields virology, 6th ed Lippincott/Williams & Wilkins, Philadelphia, PA.
-
- Griffin DE. 2013. Alphaviruses, p 651–686. In Knipe DM, Howley PM (ed), Fields virology, 6th ed Lippincott/Williams & Wilkins, Philadelphia, PA.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

