The Dengue Virus Nonstructural Protein 1 (NS1) Is Secreted from Mosquito Cells in Association with the Intracellular Cholesterol Transporter Chaperone Caveolin Complex
- PMID: 30463973
- PMCID: PMC6364000
- DOI: 10.1128/JVI.01985-18
The Dengue Virus Nonstructural Protein 1 (NS1) Is Secreted from Mosquito Cells in Association with the Intracellular Cholesterol Transporter Chaperone Caveolin Complex
Abstract
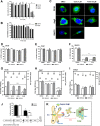
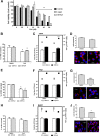
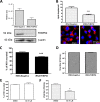
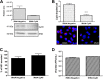
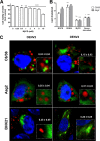
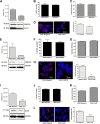
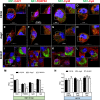
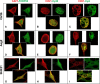
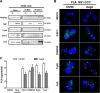
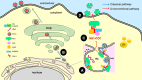
Dengue virus (DENV) is a mosquito-borne virus of the family Flaviviridae The RNA viral genome encodes three structural and seven nonstructural proteins. Nonstructural protein 1 (NS1) is a multifunctional protein actively secreted in vertebrate and mosquito cells during infection. In mosquito cells, NS1 is secreted in a caveolin-1-dependent manner by an unconventional route. The caveolin chaperone complex (CCC) is a cytoplasmic complex formed by caveolin-1 and the chaperones FKBP52, Cy40, and CyA and is responsible for the cholesterol traffic inside the cell. In this work, we demonstrate that in mosquito cells, but not in vertebrate cells, NS1 associates with and relies on the CCC for secretion. Treatment of mosquito cells with classic secretion inhibitors, such as brefeldin A, Golgicide A, and Fli-06, showed no effect on NS1 secretion but significant reductions in recombinant luciferase secretion and virion release. Silencing the expression of CAV-1 or FKBP52 with short interfering RNAs or the inhibition of CyA by cyclosporine resulted in significant decrease in NS1 secretion, again without affecting virion release. Colocalization, coimmunoprecipitation, and proximity ligation assays indicated that NS1 colocalizes and interacts with all proteins of the CCC. In addition, CAV-1 and FKBP52 expression was found augmented in DENV-infected cells. Results obtained with Zika virus-infected cells suggest that in mosquito cells, ZIKV NS1 follows the same secretory pathway as that observed for DENV NS1. These results uncover important differences in the dengue virus-cell interactions between the vertebrate host and the mosquito vector as well as novel functions for the chaperone caveolin complex.IMPORTANCE The dengue virus protein NS1 is secreted efficiently from both infected vertebrate and mosquito cells. Previously, our group reported that NS1 secretion in mosquito cells follows an unconventional secretion pathway dependent on caveolin-1. In this work, we demonstrate that in mosquito cells, but not in vertebrate cells, NS1 secretion takes place in association with the chaperone caveolin complex, a complex formed by caveolin-1 and the chaperones FKBP52, CyA, and Cy40, which are in charge of cholesterol transport inside the cell. Results obtained with ZIKV-infected mosquito cells suggest that ZIKV NS1 is released following an unconventional secretory route in association with the chaperone caveolin complex. These results uncover important differences in the virus-cell interactions between the vertebrate host and the mosquito vector, as well as novel functions for the chaperone caveolin complex. Moreover, manipulation of the NS1 secretory route may prove a valuable strategy to combat these two mosquito-borne diseases.
Keywords: NS1; Zika virus; caveolin-1; chaperone caveolin complex; dengue virus; flavivirus; mosquito cells; unconventional secretion; viral protein trafficking; yellow fever virus.
Copyright © 2019 American Society for Microbiology.
Figures



References
-
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GRW, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi: 10.1038/nature12060. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

