PAPST2 Plays Critical Roles in Removing the Stress Signaling Molecule 3'-Phosphoadenosine 5'-Phosphate from the Cytosol and Its Subsequent Degradation in Plastids and Mitochondria
- PMID: 30464037
- PMCID: PMC6391701
- DOI: 10.1105/tpc.18.00512
PAPST2 Plays Critical Roles in Removing the Stress Signaling Molecule 3'-Phosphoadenosine 5'-Phosphate from the Cytosol and Its Subsequent Degradation in Plastids and Mitochondria
Abstract
The compartmentalization of PAPS (the sulfate donor 3'-phosphoadenosine 5'-phosphosulfate) synthesis (mainly in plastids), PAPS consumption (in the cytosol), and PAP (the stress signaling molecule 3'-phosphoadenosine 5'-phosphate) degradation (in plastids and mitochondria) requires organellar transport systems for both PAPS and PAP. The plastidial transporter PAPST1 (PAPS TRANSPORTER1) delivers newly synthesized PAPS from the stroma to the cytosol. We investigated the activity of PAPST2, the closest homolog of PAPST1, which unlike PAPST1 is targeted to both the plastids and mitochondria. Biochemical characterization in Arabidopsis thaliana revealed that PAPST2 mediates the antiport of PAP, PAPS, ATP, and ADP. Strongly increased cellular PAP levels negatively affect plant growth, as observed in the fry1 papst2 mutant, which lacks the PAP-catabolizing enzyme SALT TOLERANCE 1 and PAPST2. PAP levels were specifically elevated in the cytosol of papst2 and fiery1 papst2, but not in papst1 or fry1 papst1 PAPST1 failed to complement the papst2 mutant phenotype in mitochondria, because it likely removes PAPS from the cell, as demonstrated by the increased expression of phytosulfokine genes. Overexpression of SAL1 in mitochondria rescued the phenotype of fry1 but not fry1 papst2 Therefore, PAPST2 represents an important organellar importer of PAP, providing a piece of the puzzle in our understanding of the organelle-to-nucleus PAP retrograde signaling pathway.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures





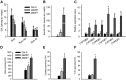

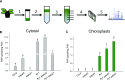


Comment in
-
Too Much, Take it Back: PAP Moves from the Cytosol to Plastids and Mitochondria for Degradation via PAPST2.Plant Cell. 2019 Jan;31(1):15-16. doi: 10.1105/tpc.18.00944. Epub 2018 Dec 20. Plant Cell. 2019. PMID: 30573470 Free PMC article. No abstract available.
References
-
- Aubry S., Smith-Unna R.D., Boursnell C.M., Kopriva S., Hibberd J.M. (2014). Transcript residency on ribosomes reveals a key role for the Arabidopsis thaliana bundle sheath in sulfur and glucosinolate metabolism. Plant J. 78: 659–673. - PubMed
-
- Berger B., Stracke R., Yatusevich R., Weisshaar B., Flügge U.I., Gigolashvili T. (2007). A simplified method for the analysis of transcription factor-promoter interactions that allows high-throughput data generation. Plant J. 50: 911–916. - PubMed
-
- Bodén M., Hawkins J. (2005). Prediction of subcellular localization using sequence-biased recurrent networks. Bioinformatics 21: 2279–2286. - PubMed
-
- Bonaventure G., Gfeller A., Rodríguez V.M., Armand F., Farmer E.E. (2007). The fou2 gain-of-function allele and the wild-type allele of Two Pore Channel 1 contribute to different extents or by different mechanisms to defense gene expression in Arabidopsis. Plant Cell Physiol. 48: 1775–1789. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

